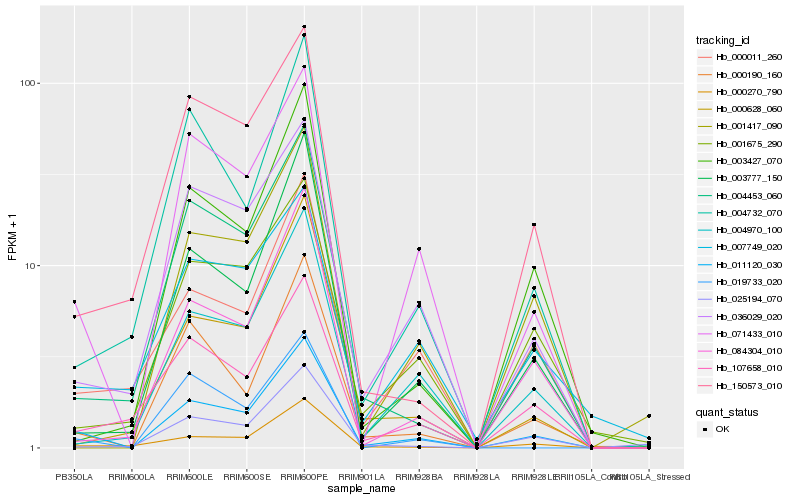
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011120_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 2 |
Hb_071433_010 |
0.125856537 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000270_790 |
0.1511345715 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 4 |
Hb_000190_160 |
0.1513048018 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000011_260 |
0.1513367921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_036029_020 |
0.1535845515 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 7 |
Hb_003777_150 |
0.1591016386 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004970_100 |
0.1600652926 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 9 |
Hb_004453_060 |
0.1609301516 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_107658_010 |
0.1629408975 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_084304_010 |
0.1678408816 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 12 |
Hb_004732_070 |
0.1747288955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001675_290 |
0.1752786957 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003427_070 |
0.1784601228 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 15 |
Hb_001417_090 |
0.1785347536 |
- |
- |
PREDICTED: uncharacterized protein LOC105650625 [Jatropha curcas] |
| 16 |
Hb_150573_010 |
0.1795193099 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 17 |
Hb_000628_060 |
0.181327618 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 18 |
Hb_025194_070 |
0.1822095501 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 19 |
Hb_019733_020 |
0.1826291266 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 20 |
Hb_007749_020 |
0.1826964905 |
- |
- |
receptor-kinase, putative [Ricinus communis] |