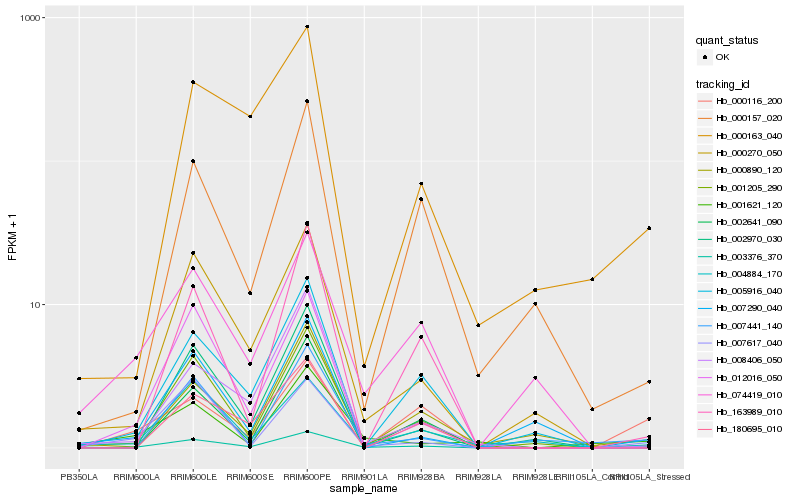
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_370 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001205_290 |
0.1863940092 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 3 |
Hb_000270_050 |
0.194003064 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 4 |
Hb_005916_040 |
0.201196113 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 5 |
Hb_012016_050 |
0.2075799676 |
- |
- |
hypothetical protein JCGZ_09036 [Jatropha curcas] |
| 6 |
Hb_007617_040 |
0.2089720398 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 7 |
Hb_004884_170 |
0.2111277709 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 8 |
Hb_000116_200 |
0.2123821279 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0005s10440g [Populus trichocarpa] |
| 9 |
Hb_008406_050 |
0.2141550075 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_001621_120 |
0.2156792201 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 11 |
Hb_163989_010 |
0.2165740758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007441_140 |
0.2166014261 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 13 |
Hb_000890_120 |
0.2186109887 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_180695_010 |
0.2218626689 |
- |
- |
hypothetical protein CICLE_v10013449mg [Citrus clementina] |
| 15 |
Hb_007290_040 |
0.2228351437 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |
| 16 |
Hb_000163_040 |
0.2231822818 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 17 |
Hb_000157_020 |
0.2232220725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002970_030 |
0.2234487859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_074419_010 |
0.2241818693 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 20 |
Hb_002641_090 |
0.2247640058 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |