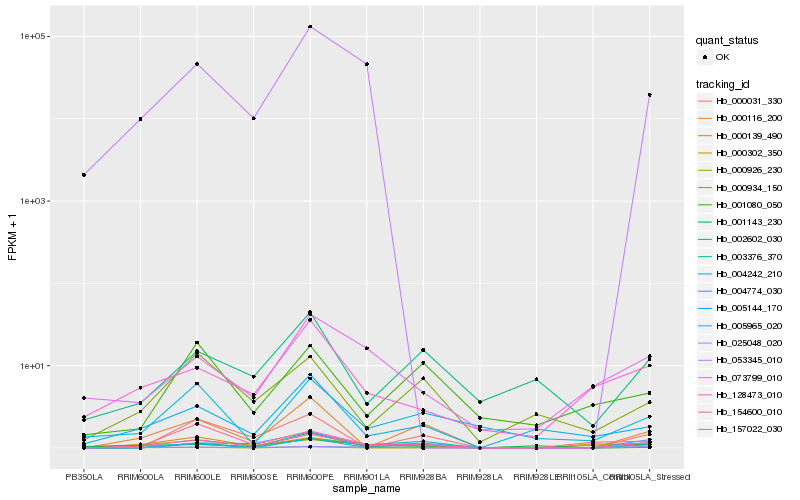
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154600_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 2 |
Hb_000116_200 |
0.2183501796 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0005s10440g [Populus trichocarpa] |
| 3 |
Hb_005965_020 |
0.2189636738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003376_370 |
0.2328046088 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000031_330 |
0.2600728894 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0157s00200g, partial [Populus trichocarpa] |
| 6 |
Hb_025048_020 |
0.2630802415 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_004774_030 |
0.2704290794 |
- |
- |
PREDICTED: uncharacterized protein LOC105648177 [Jatropha curcas] |
| 8 |
Hb_005144_170 |
0.2712397995 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000934_150 |
0.2731115277 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 10 |
Hb_000302_350 |
0.2743329526 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 11 |
Hb_004242_210 |
0.2749254571 |
- |
- |
PREDICTED: uncharacterized protein LOC105633248 [Jatropha curcas] |
| 12 |
Hb_002602_030 |
0.275498494 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000139_490 |
0.2771486431 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 14 |
Hb_000926_230 |
0.2784620253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001143_230 |
0.2799684922 |
- |
- |
beta-galactosidase family protein [Populus trichocarpa] |
| 16 |
Hb_128473_010 |
0.2836986339 |
- |
- |
- |
| 17 |
Hb_073799_010 |
0.2846745173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_053345_010 |
0.2900766853 |
- |
- |
- |
| 19 |
Hb_157022_030 |
0.298270203 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 20 |
Hb_001080_050 |
0.2996736073 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |