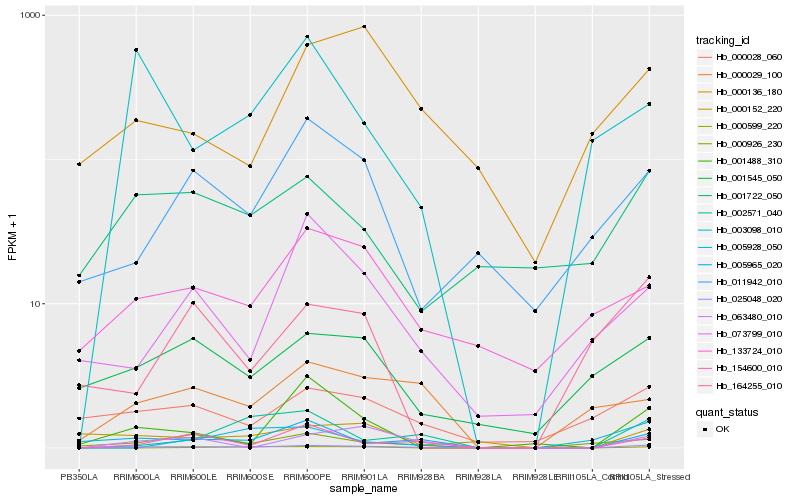
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025048_020 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_154600_010 |
0.2630802415 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 3 |
Hb_063480_010 |
0.2744079307 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like isoform X2 [Pyrus x bretschneideri] |
| 4 |
Hb_000926_230 |
0.2936027799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000029_100 |
0.2985899003 |
- |
- |
PREDICTED: nicalin-1-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000028_060 |
0.3023253979 |
- |
- |
- |
| 7 |
Hb_001545_050 |
0.3084875869 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 8 |
Hb_005965_020 |
0.3103199681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002571_040 |
0.3141543509 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 10 |
Hb_001488_310 |
0.3194788148 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_011942_010 |
0.3213833184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_133724_010 |
0.32466593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001722_050 |
0.3254641735 |
- |
- |
calcium binding family protein [Populus trichocarpa] |
| 14 |
Hb_000152_220 |
0.3255301293 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 15 |
Hb_005928_050 |
0.327671296 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_073799_010 |
0.3303847037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000136_180 |
0.3317986832 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 18 |
Hb_164255_010 |
0.3362951888 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 19 |
Hb_000599_220 |
0.3371687272 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 20 |
Hb_003098_010 |
0.3390087513 |
- |
- |
- |