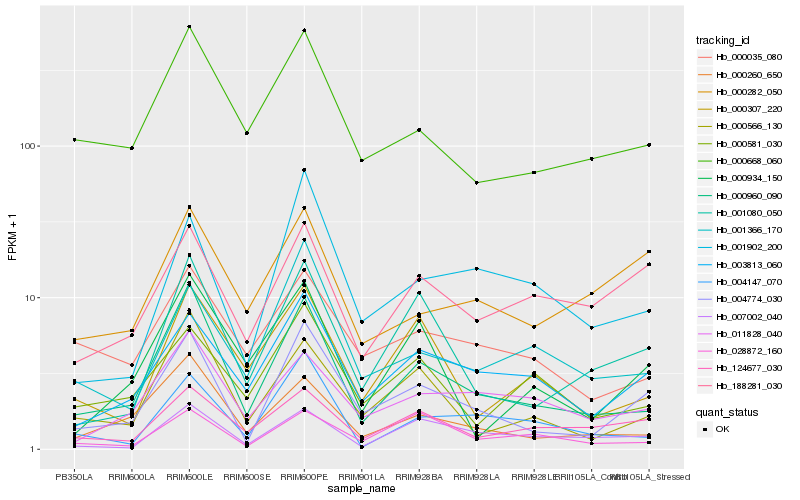
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004774_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648177 [Jatropha curcas] |
| 2 |
Hb_000960_090 |
0.1638024627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003813_060 |
0.1769678155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001080_050 |
0.1771469439 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 5 |
Hb_001902_200 |
0.194610291 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 6 |
Hb_000566_130 |
0.1976781843 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 7 |
Hb_000668_060 |
0.2054750216 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 8 |
Hb_000260_650 |
0.2055926466 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 9 |
Hb_001366_170 |
0.2108126865 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 10 |
Hb_124677_030 |
0.2187660487 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 11 |
Hb_000282_050 |
0.2199995611 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 12 |
Hb_011828_040 |
0.2207808675 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 13 |
Hb_188281_030 |
0.2220085264 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 14 |
Hb_000035_080 |
0.2258060933 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 15 |
Hb_000934_150 |
0.2295795005 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 16 |
Hb_000581_030 |
0.2297902288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004147_070 |
0.2312124671 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 18 |
Hb_007002_040 |
0.2313021361 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 19 |
Hb_000307_220 |
0.2325164848 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 20 |
Hb_028872_160 |
0.2342203043 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |