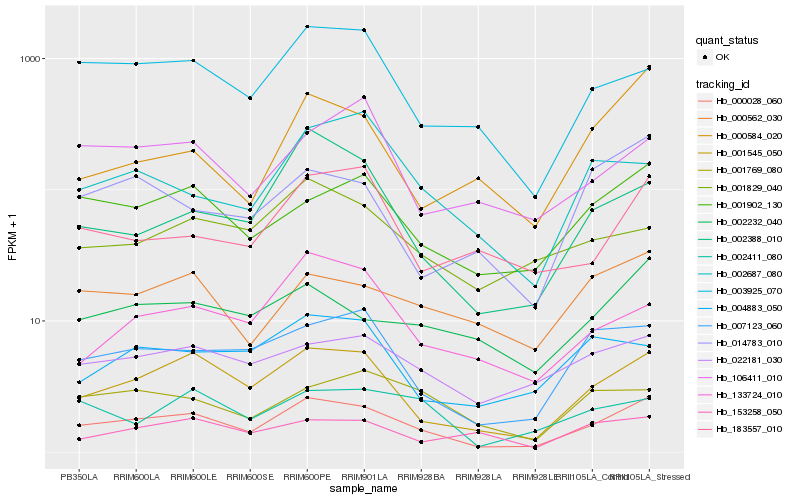
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_001545_050 |
0.1076920763 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 3 |
Hb_001902_130 |
0.1384509072 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 4 |
Hb_007123_060 |
0.1495345756 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 5 |
Hb_003925_070 |
0.1542402127 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 6 |
Hb_022181_030 |
0.1549676696 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002232_040 |
0.1592189358 |
- |
- |
- |
| 8 |
Hb_000562_030 |
0.161777037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002411_080 |
0.1755341562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_133724_010 |
0.1772739955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002388_010 |
0.1773075763 |
- |
- |
- |
| 12 |
Hb_014783_010 |
0.1775489578 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001769_080 |
0.1780771002 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |
| 14 |
Hb_001829_040 |
0.1786452673 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 15 |
Hb_183557_010 |
0.1791153731 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 16 |
Hb_004883_050 |
0.1821371249 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 17 |
Hb_106411_010 |
0.1832391318 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 18 |
Hb_153258_050 |
0.1839531823 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 19 |
Hb_002687_080 |
0.1849712559 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 20 |
Hb_000584_020 |
0.1875900309 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |