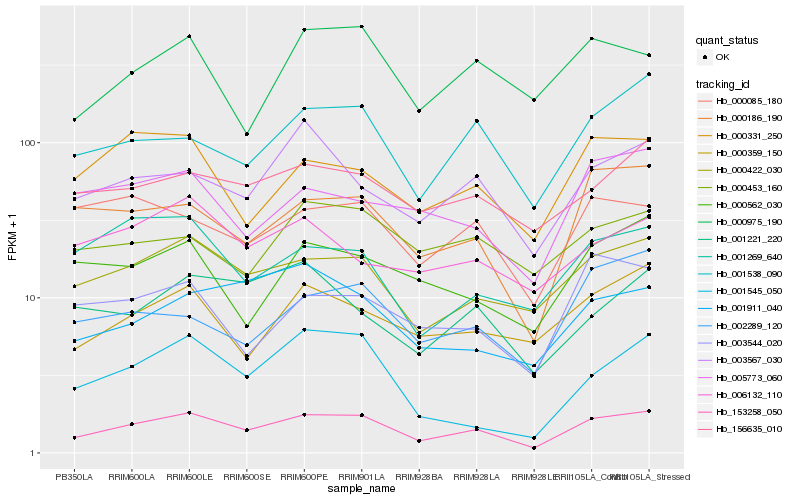
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153258_050 |
0.0 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 2 |
Hb_000422_030 |
0.1076375928 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 3 |
Hb_001538_090 |
0.1267393593 |
- |
- |
40S ribosomal protein S29 [Morus notabilis] |
| 4 |
Hb_000186_190 |
0.1297295243 |
- |
- |
PREDICTED: uncharacterized protein At4g22160 [Jatropha curcas] |
| 5 |
Hb_156635_010 |
0.1362460397 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 6 |
Hb_000359_150 |
0.1376498894 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003567_030 |
0.138455434 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000331_250 |
0.1431172322 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 9 |
Hb_000975_190 |
0.1452123257 |
- |
- |
histone H4 [Zea mays] |
| 10 |
Hb_000085_180 |
0.1473879646 |
- |
- |
PREDICTED: syntaxin-52-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001911_040 |
0.1474968323 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 12 |
Hb_000562_030 |
0.1497975063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005773_060 |
0.1503908983 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 14 |
Hb_002289_120 |
0.1505574847 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000453_160 |
0.1511175473 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 16 |
Hb_001221_220 |
0.1517525255 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 17 |
Hb_003544_020 |
0.1527359503 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 18 |
Hb_001269_640 |
0.1530895197 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |
| 19 |
Hb_001545_050 |
0.1533633723 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 20 |
Hb_006132_110 |
0.1554801314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |