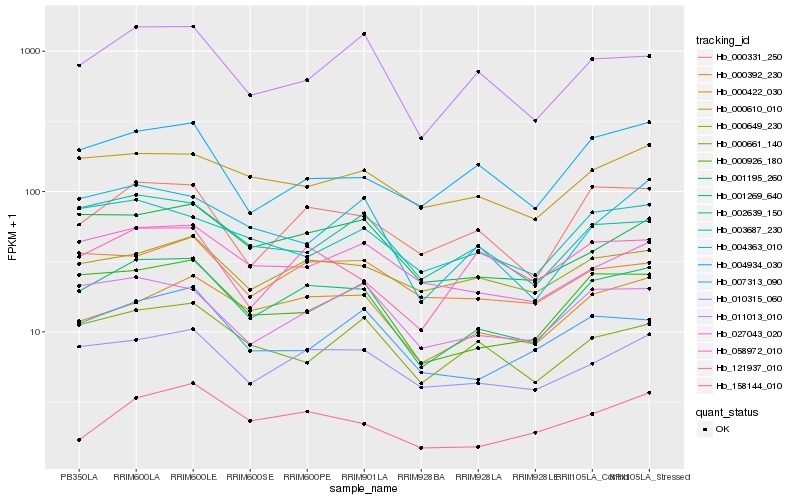
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_640 |
0.0 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |
| 2 |
Hb_000926_180 |
0.0850078164 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 3 |
Hb_010315_060 |
0.0891547483 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 4 |
Hb_000331_250 |
0.0908008263 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 5 |
Hb_000422_030 |
0.100279321 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 6 |
Hb_027043_020 |
0.1040332627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001195_260 |
0.1049163443 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 8 |
Hb_011013_010 |
0.1075553351 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 9 |
Hb_002639_150 |
0.1115273812 |
- |
- |
- |
| 10 |
Hb_007313_090 |
0.1119073892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000392_230 |
0.1135327094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000610_010 |
0.1138669057 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_004934_030 |
0.1150974116 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 14 |
Hb_058972_010 |
0.115263362 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000649_230 |
0.1152842602 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 16 |
Hb_121937_010 |
0.1184341807 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 17 |
Hb_004363_010 |
0.1206875546 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 18 |
Hb_003687_230 |
0.1208185512 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 19 |
Hb_158144_010 |
0.1210039805 |
- |
- |
- |
| 20 |
Hb_000661_140 |
0.121176156 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |