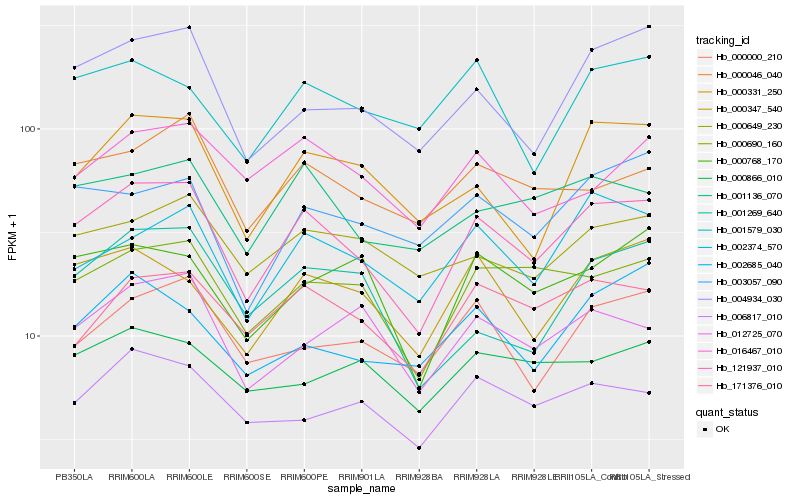
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121937_010 |
0.0 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 2 |
Hb_000690_160 |
0.093328536 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 3 |
Hb_000347_540 |
0.100866337 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_001136_070 |
0.1010916525 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 5 |
Hb_171376_010 |
0.1012516692 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004934_030 |
0.1022774302 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 7 |
Hb_002374_570 |
0.1040652791 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 8 |
Hb_000046_040 |
0.1056956248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000331_250 |
0.1062056588 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 10 |
Hb_006817_010 |
0.1113599707 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000000_210 |
0.1114296029 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000649_230 |
0.1118440056 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 13 |
Hb_012725_070 |
0.1123131011 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000768_170 |
0.1132819686 |
- |
- |
hypothetical protein JCGZ_02624 [Jatropha curcas] |
| 15 |
Hb_003057_090 |
0.1147818558 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 16 |
Hb_016467_010 |
0.1161470204 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 17 |
Hb_001269_640 |
0.1184341807 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |
| 18 |
Hb_001579_030 |
0.118957064 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 19 |
Hb_002685_040 |
0.1222428157 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 20 |
Hb_000866_010 |
0.1236211107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |