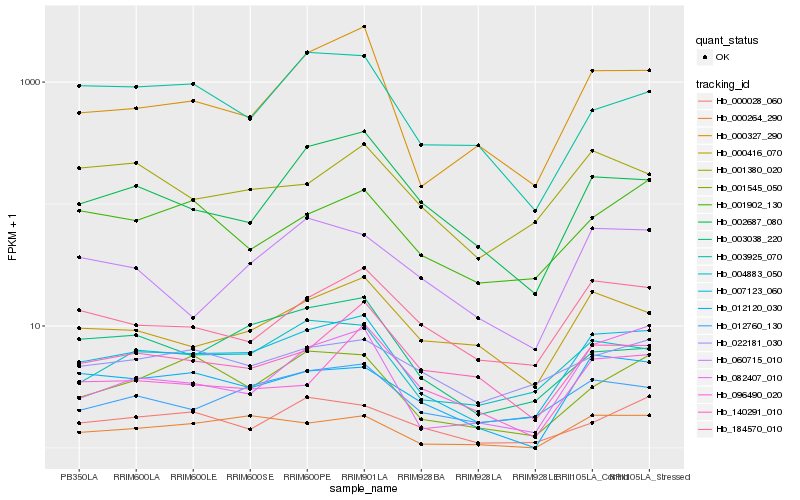
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007123_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 2 |
Hb_004883_050 |
0.1147541759 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 3 |
Hb_012760_130 |
0.1306169596 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 4 |
Hb_001545_050 |
0.1408703992 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 5 |
Hb_000327_290 |
0.1416009924 |
- |
- |
Histone H4 [Triticum urartu] |
| 6 |
Hb_184570_010 |
0.1429443187 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 7 |
Hb_012120_030 |
0.1459816291 |
- |
- |
- |
| 8 |
Hb_003038_220 |
0.1469253856 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 9 |
Hb_000264_290 |
0.1469778427 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 10 |
Hb_000416_070 |
0.1490400246 |
- |
- |
hypothetical protein JCGZ_21620 [Jatropha curcas] |
| 11 |
Hb_000028_060 |
0.1495345756 |
- |
- |
- |
| 12 |
Hb_002687_080 |
0.1522174352 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 13 |
Hb_022181_030 |
0.1522847691 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 14 |
Hb_082407_010 |
0.1540134042 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 15 |
Hb_001902_130 |
0.1556528357 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 16 |
Hb_001380_020 |
0.1587230439 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 17 |
Hb_140291_010 |
0.1597379803 |
- |
- |
hypothetical protein JCGZ_17222 [Jatropha curcas] |
| 18 |
Hb_003925_070 |
0.1613840002 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 19 |
Hb_060715_010 |
0.1615594067 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 20 |
Hb_096490_020 |
0.1623176193 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |