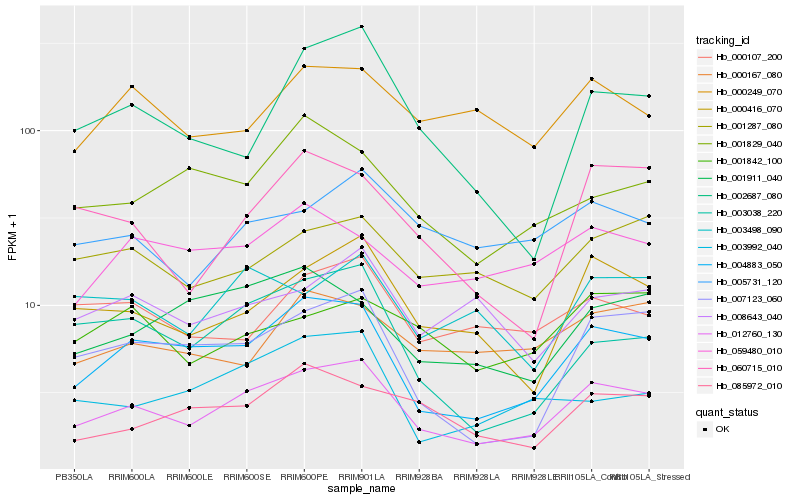
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 2 |
Hb_004883_050 |
0.0926983936 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 3 |
Hb_000416_070 |
0.1127995504 |
- |
- |
hypothetical protein JCGZ_21620 [Jatropha curcas] |
| 4 |
Hb_007123_060 |
0.1306169596 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 5 |
Hb_005731_120 |
0.139136066 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20 [Jatropha curcas] |
| 6 |
Hb_001287_080 |
0.1419011865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003498_090 |
0.1435950241 |
- |
- |
hypothetical protein RCOM_0867850 [Ricinus communis] |
| 8 |
Hb_085972_010 |
0.1445518242 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 9 |
Hb_003038_220 |
0.1445520965 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 10 |
Hb_000107_200 |
0.1473665247 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |
| 11 |
Hb_001911_040 |
0.1484192911 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 12 |
Hb_059480_010 |
0.1488337987 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_008643_040 |
0.1489674006 |
- |
- |
PREDICTED: jacalin-related lectin 3-like [Jatropha curcas] |
| 14 |
Hb_060715_010 |
0.1495920061 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 15 |
Hb_003992_040 |
0.1496478448 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 16 |
Hb_000249_070 |
0.1505997421 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 17 |
Hb_001842_100 |
0.1507087942 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 18 |
Hb_002687_080 |
0.1509708318 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 19 |
Hb_000167_080 |
0.1516072496 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 20 |
Hb_001829_040 |
0.1519390672 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |