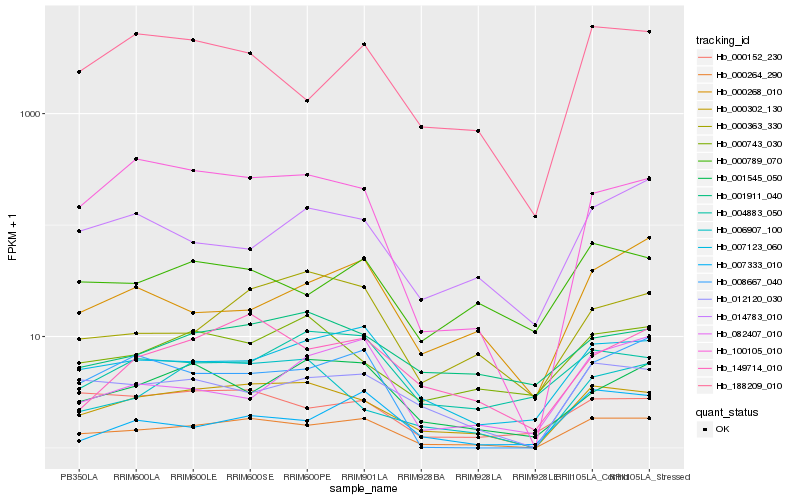
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_290 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 2 |
Hb_000302_130 |
0.1202001146 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 3 |
Hb_007123_060 |
0.1469778427 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 4 |
Hb_012120_030 |
0.1544617499 |
- |
- |
- |
| 5 |
Hb_008667_040 |
0.1750674674 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 6 |
Hb_188209_010 |
0.1753689582 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_149714_010 |
0.1803194403 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 8 |
Hb_100105_010 |
0.1835711326 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 9 |
Hb_000789_070 |
0.1878024673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000152_230 |
0.1890369991 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 11 |
Hb_000363_330 |
0.1911143309 |
- |
- |
- |
| 12 |
Hb_001545_050 |
0.195292746 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 13 |
Hb_007333_010 |
0.2058986604 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_004883_050 |
0.2061409054 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 15 |
Hb_006907_100 |
0.2074470497 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 16 |
Hb_000268_010 |
0.208888122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001911_040 |
0.2101632827 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 18 |
Hb_014783_010 |
0.2126687361 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000743_030 |
0.212807611 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 20 |
Hb_082407_010 |
0.2130438693 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |