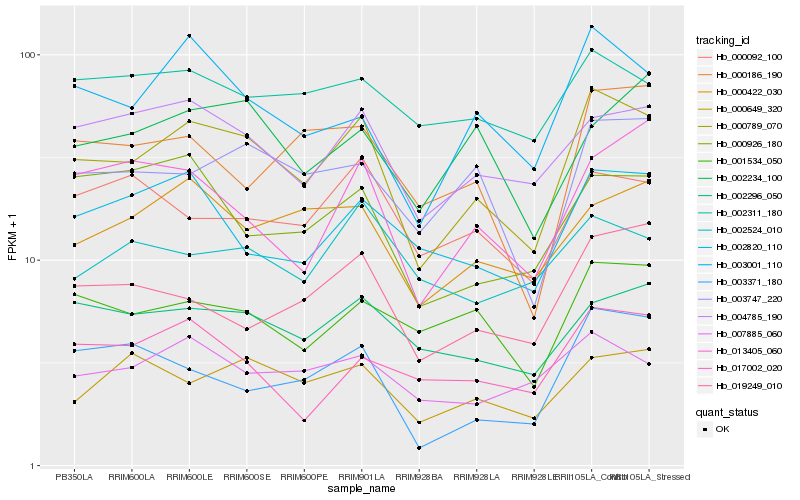
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003001_110 |
0.1203518557 |
- |
- |
oleoyl-acyl carrier protein thioesterase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004785_190 |
0.1223222707 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 4 |
Hb_003747_220 |
0.1268704382 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002296_050 |
0.1307776923 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 6 |
Hb_000422_030 |
0.1320272115 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 7 |
Hb_001534_050 |
0.1343416722 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 8 |
Hb_002820_110 |
0.1352200486 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 9 |
Hb_007885_060 |
0.1363572065 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 10 |
Hb_000649_320 |
0.1375817201 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 11 |
Hb_019249_010 |
0.1375836869 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 12 |
Hb_000092_100 |
0.1415719637 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 13 |
Hb_002311_180 |
0.1434593412 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 14 |
Hb_000186_190 |
0.144552344 |
- |
- |
PREDICTED: uncharacterized protein At4g22160 [Jatropha curcas] |
| 15 |
Hb_013405_060 |
0.1449581849 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000926_180 |
0.1465602278 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 17 |
Hb_003371_180 |
0.1483194245 |
- |
- |
- |
| 18 |
Hb_002524_010 |
0.149875836 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 19 |
Hb_002234_100 |
0.1499955302 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 20 |
Hb_017002_020 |
0.1502365068 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |