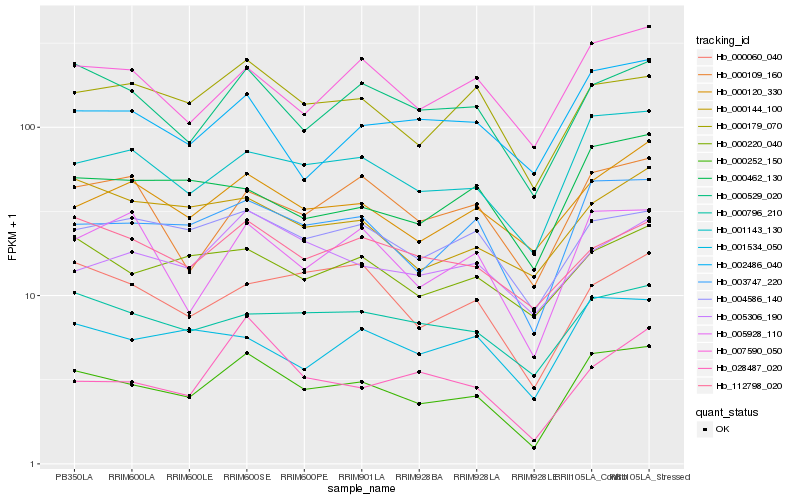
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003747_220 |
0.0812662 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001143_130 |
0.0902167398 |
- |
- |
peroxidase 2 [Hevea brasiliensis] |
| 4 |
Hb_000529_020 |
0.1063471529 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 5 |
Hb_007590_050 |
0.1157013987 |
- |
- |
NudC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000179_070 |
0.1178237939 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 7 |
Hb_005928_110 |
0.1181472068 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 8 |
Hb_000120_330 |
0.1208755616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000796_210 |
0.1223437552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004586_140 |
0.1248303421 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 11 |
Hb_000220_040 |
0.1258242906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001534_050 |
0.1260729304 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 13 |
Hb_000462_130 |
0.1265895108 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 14 |
Hb_000109_160 |
0.1287209711 |
- |
- |
hypothetical protein PRUPE_ppa012786mg [Prunus persica] |
| 15 |
Hb_000060_040 |
0.1305290221 |
- |
- |
hypothetical protein JCGZ_21620 [Jatropha curcas] |
| 16 |
Hb_002486_040 |
0.1307570634 |
- |
- |
- |
| 17 |
Hb_000144_100 |
0.1308396416 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_112798_020 |
0.1311288387 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 19 |
Hb_005306_190 |
0.1329237152 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 20 |
Hb_028487_020 |
0.1334160944 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |