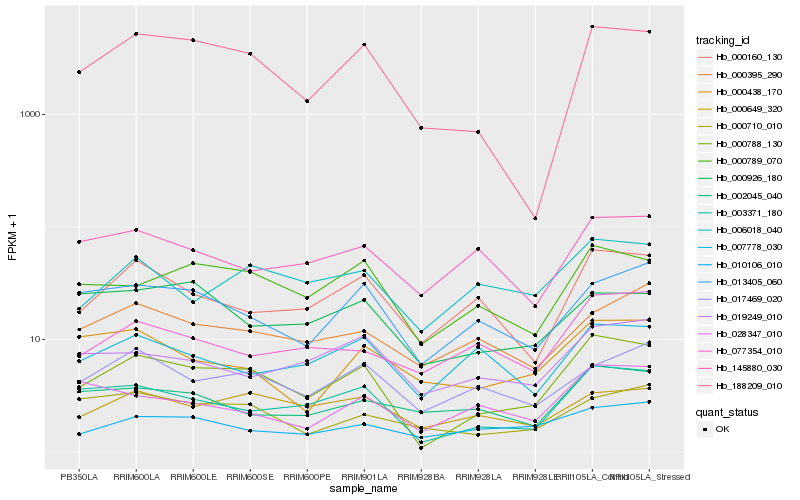
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000788_130 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 2 |
Hb_003371_180 |
0.1243167396 |
- |
- |
- |
| 3 |
Hb_188209_010 |
0.1511414181 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000789_070 |
0.1536222505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000160_130 |
0.1664068754 |
- |
- |
PREDICTED: uncharacterized protein LOC105648215 [Jatropha curcas] |
| 6 |
Hb_013405_060 |
0.1725792807 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006018_040 |
0.1761889187 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 8 |
Hb_017469_020 |
0.1825395641 |
- |
- |
- |
| 9 |
Hb_019249_010 |
0.1833515743 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 10 |
Hb_000649_320 |
0.1841180322 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 11 |
Hb_010106_010 |
0.1883380955 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 12 |
Hb_077354_010 |
0.1897963154 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 13 |
Hb_000710_010 |
0.1911825259 |
- |
- |
PREDICTED: uncharacterized protein LOC105630356 [Jatropha curcas] |
| 14 |
Hb_007778_030 |
0.192478412 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000395_290 |
0.1937009365 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 16 |
Hb_000438_170 |
0.1937505912 |
- |
- |
- |
| 17 |
Hb_145880_030 |
0.1951216739 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 18 |
Hb_000926_180 |
0.1951764526 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 19 |
Hb_028347_010 |
0.1956325533 |
- |
- |
- |
| 20 |
Hb_002045_040 |
0.197439612 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |