| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
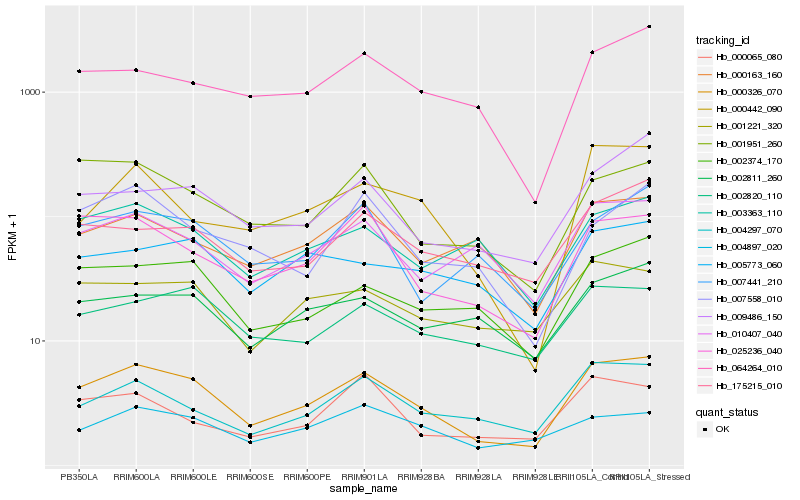
Hb_000326_070 |
0.0 |
- |
- |
phenylalanyl-tRNA synthetase beta chain, putative [Ricinus communis] |
| 2 |
Hb_025236_040 |
0.1247328271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002374_170 |
0.131243449 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 4 |
Hb_004297_070 |
0.1317398327 |
- |
- |
hypothetical protein POPTR_0009s14820g [Populus trichocarpa] |
| 5 |
Hb_001951_260 |
0.1345483185 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 6 |
Hb_064264_010 |
0.1398457531 |
- |
- |
hypothetical protein [Zea mays] |
| 7 |
Hb_175215_010 |
0.1402134736 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 8 |
Hb_002820_110 |
0.1417006771 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 9 |
Hb_009486_150 |
0.1418344981 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 10 |
Hb_010407_040 |
0.1432878693 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 11 |
Hb_001221_320 |
0.1461591676 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000442_090 |
0.1465177317 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 13 |
Hb_007441_210 |
0.1466086754 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 14 |
Hb_004897_020 |
0.1493765209 |
- |
- |
PREDICTED: (-)-germacrene D synthase-like [Populus euphratica] |
| 15 |
Hb_002811_260 |
0.1504577715 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 16 |
Hb_007558_010 |
0.1504610303 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 17 |
Hb_000065_080 |
0.1505962575 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, chloroplastic-like [Nelumbo nucifera] |
| 18 |
Hb_003363_110 |
0.1509084826 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000163_160 |
0.1512419173 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 20 |
Hb_005773_060 |
0.1516448925 |
- |
- |
defender against cell death, putative [Ricinus communis] |