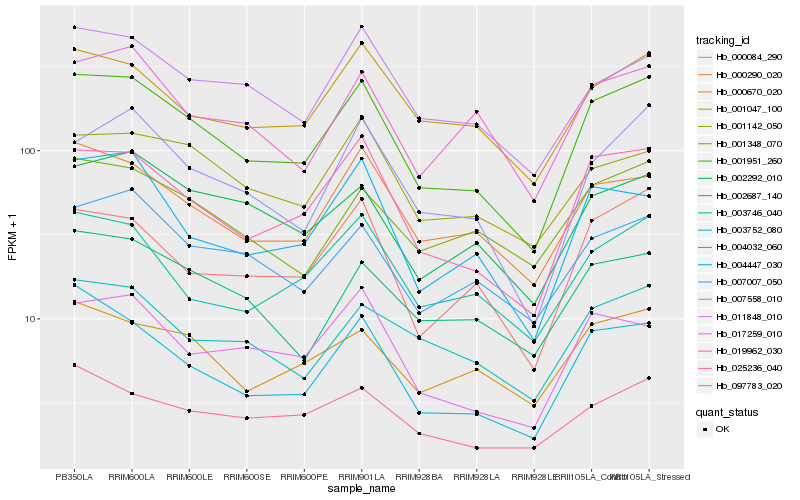
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_260 |
0.0 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 2 |
Hb_025236_040 |
0.0599349397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000290_020 |
0.0881304792 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 4 |
Hb_004032_060 |
0.090430666 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 5 |
Hb_000084_290 |
0.0906996735 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 6 |
Hb_007558_010 |
0.094121532 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 7 |
Hb_001047_100 |
0.0980830098 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 8 |
Hb_004447_030 |
0.0992460797 |
- |
- |
PREDICTED: glucose-induced degradation protein 4 homolog [Jatropha curcas] |
| 9 |
Hb_007007_050 |
0.0993670993 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 10 |
Hb_003746_040 |
0.1000579466 |
- |
- |
PREDICTED: protein N-terminal glutamine amidohydrolase [Jatropha curcas] |
| 11 |
Hb_000670_020 |
0.1007643335 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 12 |
Hb_011848_010 |
0.1008545375 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 13 |
Hb_003752_080 |
0.1019749782 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002687_140 |
0.1022974869 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 15 |
Hb_017259_010 |
0.1023918892 |
- |
- |
- |
| 16 |
Hb_002292_010 |
0.1027127715 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 17 |
Hb_001348_070 |
0.104649092 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_019962_030 |
0.1062739044 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 19 |
Hb_097783_020 |
0.1067495788 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 20 |
Hb_001142_050 |
0.1075682085 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |