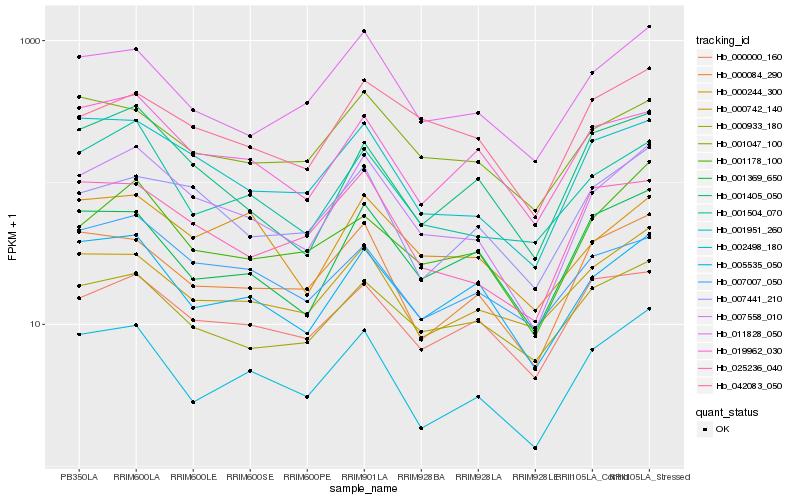
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007558_010 |
0.0 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 2 |
Hb_001951_260 |
0.094121532 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 3 |
Hb_005535_050 |
0.106322989 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001504_070 |
0.1083649529 |
- |
- |
PREDICTED: 60S ribosomal protein L7-1 [Jatropha curcas] |
| 5 |
Hb_011828_050 |
0.1140132539 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 6 |
Hb_007441_210 |
0.115933126 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 7 |
Hb_019962_030 |
0.1182108895 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 8 |
Hb_000084_290 |
0.1183256526 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 9 |
Hb_042083_050 |
0.1188136384 |
- |
- |
60S ribosomal protein L11, putative [Ricinus communis] |
| 10 |
Hb_001178_100 |
0.1196510279 |
- |
- |
60S acidic ribosomal protein P2-4 [Theobroma cacao] |
| 11 |
Hb_000933_180 |
0.1209783796 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_025236_040 |
0.1210956551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000742_140 |
0.1217214416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001405_050 |
0.1223284502 |
- |
- |
PREDICTED: uncharacterized protein LOC100500641 [Glycine max] |
| 15 |
Hb_007007_050 |
0.1239550151 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 16 |
Hb_000000_160 |
0.1239789248 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 17 |
Hb_002498_180 |
0.1240362416 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 18 |
Hb_001369_650 |
0.1244379474 |
- |
- |
PREDICTED: chloride conductance regulatory protein ICln [Jatropha curcas] |
| 19 |
Hb_000244_300 |
0.1245896955 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001047_100 |
0.1251726833 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |