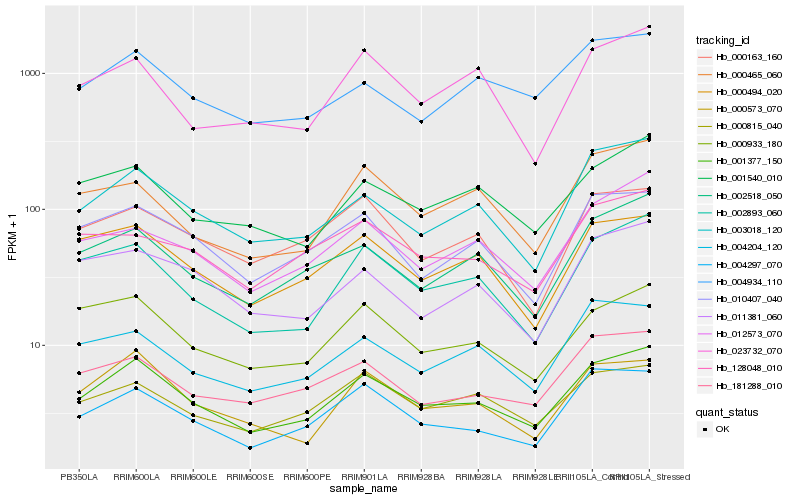
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_14266 [Jatropha curcas] |
| 2 |
Hb_002893_060 |
0.0782705547 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 3 |
Hb_004297_070 |
0.0814819138 |
- |
- |
hypothetical protein POPTR_0009s14820g [Populus trichocarpa] |
| 4 |
Hb_003018_120 |
0.0832966492 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 5 |
Hb_000573_070 |
0.0841722059 |
- |
- |
hypothetical protein JCGZ_14808 [Jatropha curcas] |
| 6 |
Hb_002518_050 |
0.0847334313 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 7 |
Hb_000933_180 |
0.088497102 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000494_020 |
0.0888711185 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 5A [Jatropha curcas] |
| 9 |
Hb_010407_040 |
0.0905032203 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 10 |
Hb_001540_010 |
0.0922449882 |
- |
- |
PREDICTED: uncharacterized protein LOC105639920 [Jatropha curcas] |
| 11 |
Hb_000815_040 |
0.0932317438 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_15623 [Jatropha curcas] |
| 12 |
Hb_181288_010 |
0.0933855508 |
- |
- |
rac-GTP binding protein, putative [Ricinus communis] |
| 13 |
Hb_012573_070 |
0.0987464463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004204_120 |
0.0997396677 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 15 |
Hb_004934_110 |
0.101696387 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 16 |
Hb_011381_060 |
0.1021876575 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 17 |
Hb_000465_060 |
0.1034226655 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_023732_070 |
0.1044184183 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 19 |
Hb_128048_010 |
0.1059429081 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 20 |
Hb_000163_160 |
0.1061279172 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |