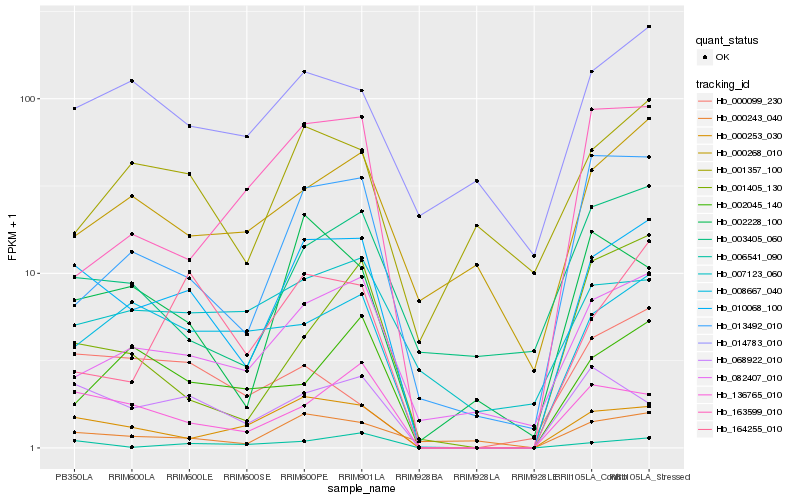
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010068_100 |
0.0 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000253_030 |
0.1666538916 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 3 |
Hb_136765_010 |
0.1827127692 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_068922_010 |
0.1972703697 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 5 |
Hb_006541_090 |
0.1990828538 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 6 |
Hb_082407_010 |
0.2003927243 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_008667_040 |
0.2024215399 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 8 |
Hb_000243_040 |
0.2045331189 |
- |
- |
- |
| 9 |
Hb_013492_010 |
0.2065817968 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 10 |
Hb_002228_100 |
0.2136821461 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_164255_010 |
0.2155964109 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 12 |
Hb_014783_010 |
0.2344130747 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_002045_140 |
0.2383575915 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 14 |
Hb_003405_060 |
0.239056044 |
- |
- |
- |
| 15 |
Hb_163599_010 |
0.2405608337 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 16 |
Hb_007123_060 |
0.2409718354 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 17 |
Hb_000268_010 |
0.2428778556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000099_230 |
0.2435369519 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 19 |
Hb_001405_130 |
0.2445367345 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 20 |
Hb_001357_100 |
0.2457744195 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |