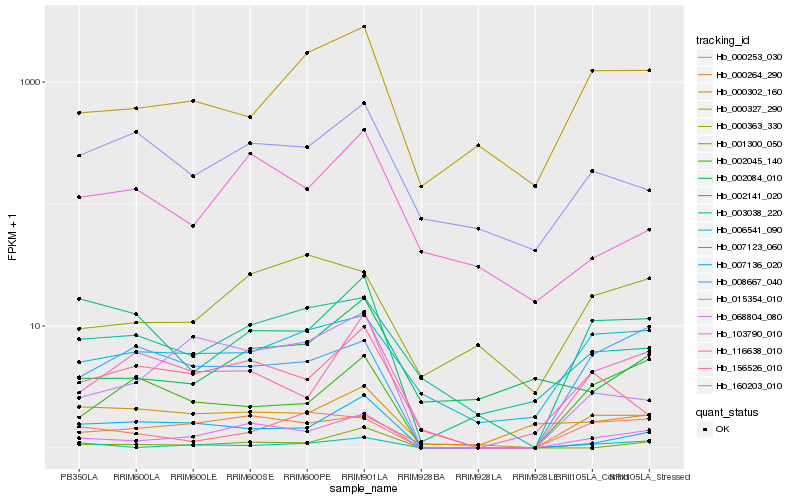
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103790_010 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 2 |
Hb_068804_080 |
0.2175671034 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 3 |
Hb_006541_090 |
0.2225056369 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 4 |
Hb_000264_290 |
0.2270985514 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 5 |
Hb_160203_010 |
0.2361077236 |
- |
- |
- |
| 6 |
Hb_007136_020 |
0.2393631932 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 7 |
Hb_001300_050 |
0.2412360196 |
- |
- |
- |
| 8 |
Hb_156526_010 |
0.2421958674 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 9 |
Hb_008667_040 |
0.2506242175 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 10 |
Hb_002141_020 |
0.2514041788 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 11 |
Hb_003038_220 |
0.2515862429 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 12 |
Hb_116638_010 |
0.2542521036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007123_060 |
0.2545575311 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 14 |
Hb_000363_330 |
0.2561760659 |
- |
- |
- |
| 15 |
Hb_000253_030 |
0.2622062072 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 16 |
Hb_000302_160 |
0.2667227899 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 17 |
Hb_002045_140 |
0.2671838542 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 18 |
Hb_000327_290 |
0.2698033462 |
- |
- |
Histone H4 [Triticum urartu] |
| 19 |
Hb_002084_010 |
0.2711015693 |
- |
- |
hypothetical protein VITISV_017990 [Vitis vinifera] |
| 20 |
Hb_015354_010 |
0.2797955886 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |