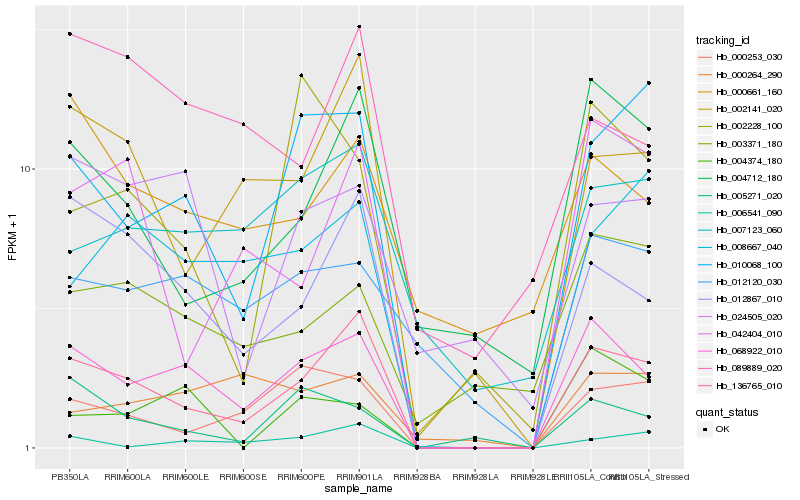
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068922_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 2 |
Hb_136765_010 |
0.1524169906 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_012867_010 |
0.1880539536 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 4 |
Hb_010068_100 |
0.1972703697 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 5 |
Hb_012120_030 |
0.2192707314 |
- |
- |
- |
| 6 |
Hb_000253_030 |
0.2237222873 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 7 |
Hb_005271_020 |
0.2300682178 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 8 |
Hb_002141_020 |
0.2322355982 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 9 |
Hb_004374_180 |
0.2376472156 |
- |
- |
GMP synthase, putative [Ricinus communis] |
| 10 |
Hb_008667_040 |
0.2390977281 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 11 |
Hb_024505_020 |
0.2393333037 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 12 |
Hb_004712_180 |
0.2419758018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007123_060 |
0.2430765526 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 14 |
Hb_002228_100 |
0.2434644522 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_003371_180 |
0.2457158994 |
- |
- |
- |
| 16 |
Hb_000661_160 |
0.2461328849 |
- |
- |
hexokinase [Manihot esculenta] |
| 17 |
Hb_000264_290 |
0.2484754665 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 18 |
Hb_089889_020 |
0.2528405515 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 19 |
Hb_006541_090 |
0.255931503 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 20 |
Hb_042404_010 |
0.2563978384 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |