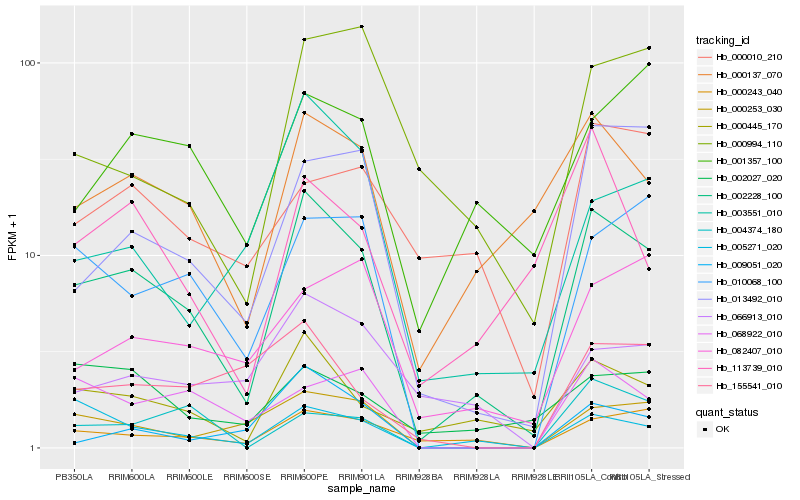
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002228_100 |
0.0 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 2 |
Hb_000445_170 |
0.1780217997 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 3 |
Hb_000137_070 |
0.1903161592 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000243_040 |
0.1980828538 |
- |
- |
- |
| 5 |
Hb_013492_010 |
0.2081354702 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 6 |
Hb_010068_100 |
0.2136821461 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005271_020 |
0.216205901 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 8 |
Hb_000253_030 |
0.2198177117 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 9 |
Hb_003551_010 |
0.2392928189 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 10 |
Hb_113739_010 |
0.2396425123 |
- |
- |
hypothetical protein JCGZ_06379 [Jatropha curcas] |
| 11 |
Hb_001357_100 |
0.2406559675 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 12 |
Hb_066913_010 |
0.2412850569 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 13 |
Hb_082407_010 |
0.2415229623 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 14 |
Hb_000994_110 |
0.2434310123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_068922_010 |
0.2434644522 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 16 |
Hb_009051_020 |
0.2461441008 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 17 |
Hb_004374_180 |
0.2475337614 |
- |
- |
GMP synthase, putative [Ricinus communis] |
| 18 |
Hb_000010_210 |
0.2519205078 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 19 |
Hb_002027_020 |
0.2527376878 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 20 |
Hb_155541_010 |
0.254488404 |
- |
- |
- |