| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
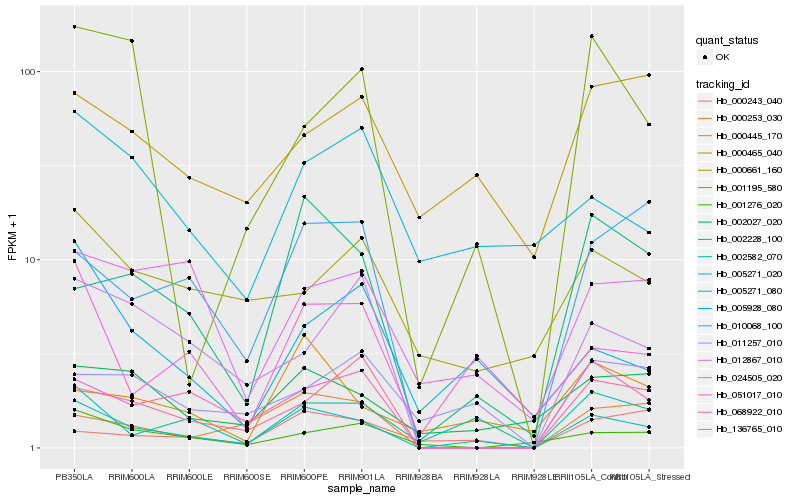
Hb_005271_020 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 2 |
Hb_002582_070 |
0.1977012851 |
- |
- |
- |
| 3 |
Hb_051017_010 |
0.2160102199 |
- |
- |
Uncharacterized protein TCM_031047 [Theobroma cacao] |
| 4 |
Hb_002228_100 |
0.216205901 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_002027_020 |
0.2248644382 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 6 |
Hb_068922_010 |
0.2300682178 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 7 |
Hb_024505_020 |
0.2419459572 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 8 |
Hb_000445_170 |
0.2467986109 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 9 |
Hb_005928_080 |
0.2474432459 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000253_030 |
0.250177002 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 11 |
Hb_010068_100 |
0.2506098867 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 12 |
Hb_136765_010 |
0.252816186 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001276_020 |
0.257894286 |
- |
- |
hypothetical protein VITISV_003263 [Vitis vinifera] |
| 14 |
Hb_001195_580 |
0.2605468851 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 15 |
Hb_012867_010 |
0.2610273914 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 16 |
Hb_000661_160 |
0.2612662556 |
- |
- |
hexokinase [Manihot esculenta] |
| 17 |
Hb_011257_010 |
0.2613394605 |
- |
- |
- |
| 18 |
Hb_000243_040 |
0.2617556801 |
- |
- |
- |
| 19 |
Hb_000465_040 |
0.2620520772 |
- |
- |
PREDICTED: uncharacterized protein LOC103433005 isoform X1 [Malus domestica] |
| 20 |
Hb_005271_080 |
0.2624814777 |
- |
- |
receptor kinase, putative [Ricinus communis] |