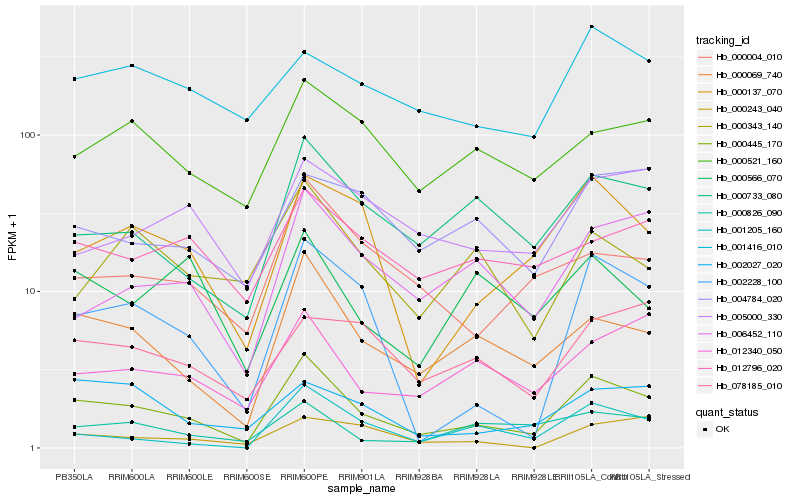
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_170 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 2 |
Hb_000069_740 |
0.1443404189 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 3 |
Hb_002228_100 |
0.1780217997 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_000137_070 |
0.179378745 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000733_080 |
0.1823800657 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 6 |
Hb_006452_110 |
0.1932359754 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 7 |
Hb_012340_050 |
0.1954134319 |
- |
- |
PREDICTED: actin-related protein 5 [Jatropha curcas] |
| 8 |
Hb_000004_010 |
0.2008371102 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 9 |
Hb_001205_160 |
0.201577584 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 10 |
Hb_000521_160 |
0.2024753363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000826_090 |
0.2026325668 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 12 |
Hb_002027_020 |
0.2075228853 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 13 |
Hb_001416_010 |
0.2158315309 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 14 |
Hb_078185_010 |
0.216185744 |
- |
- |
- |
| 15 |
Hb_004784_020 |
0.2163193519 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 16 |
Hb_005000_330 |
0.2179646273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000243_040 |
0.2180237241 |
- |
- |
- |
| 18 |
Hb_012796_020 |
0.2182033936 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 19 |
Hb_000343_140 |
0.2184100624 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 20 |
Hb_000566_070 |
0.218696355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |