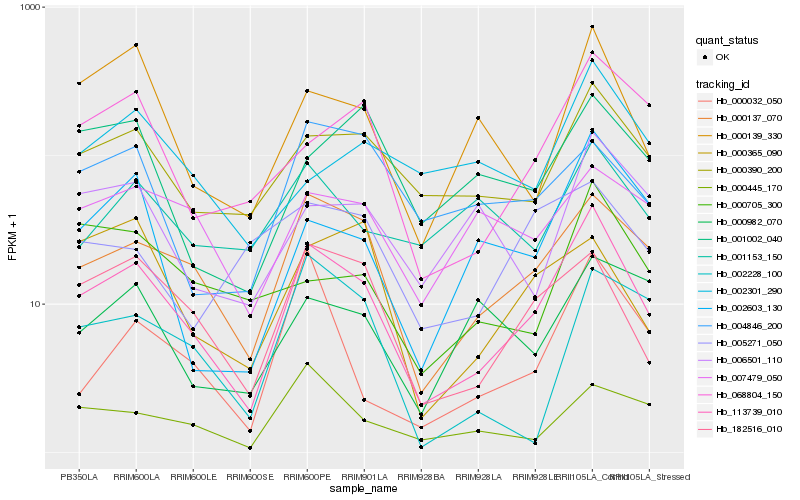
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113739_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06379 [Jatropha curcas] |
| 2 |
Hb_000137_070 |
0.1531494058 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 3 |
Hb_182516_010 |
0.1811596768 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 4 |
Hb_000390_200 |
0.184963854 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 5 |
Hb_068804_150 |
0.204116063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000139_330 |
0.206421485 |
- |
- |
short chain dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002603_130 |
0.2096114525 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 8 |
Hb_000445_170 |
0.2267213181 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 9 |
Hb_006501_110 |
0.2297894728 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000032_050 |
0.2319632833 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 11 |
Hb_000705_300 |
0.2343361654 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 12 |
Hb_007479_050 |
0.2350409932 |
- |
- |
PREDICTED: uncharacterized protein LOC105649412 [Jatropha curcas] |
| 13 |
Hb_004846_200 |
0.2372680209 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 14 |
Hb_001153_150 |
0.2374121412 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 15 |
Hb_002228_100 |
0.2396425123 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_001002_040 |
0.240465453 |
- |
- |
PREDICTED: ATP-citrate synthase beta chain protein 2 [Gossypium raimondii] |
| 17 |
Hb_000365_090 |
0.2444441492 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_002301_290 |
0.2453006208 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 19 |
Hb_005271_050 |
0.2477767539 |
- |
- |
Tubulin beta-8 chain -like protein [Gossypium arboreum] |
| 20 |
Hb_000982_070 |
0.2486105801 |
- |
- |
unnamed protein product [Vitis vinifera] |