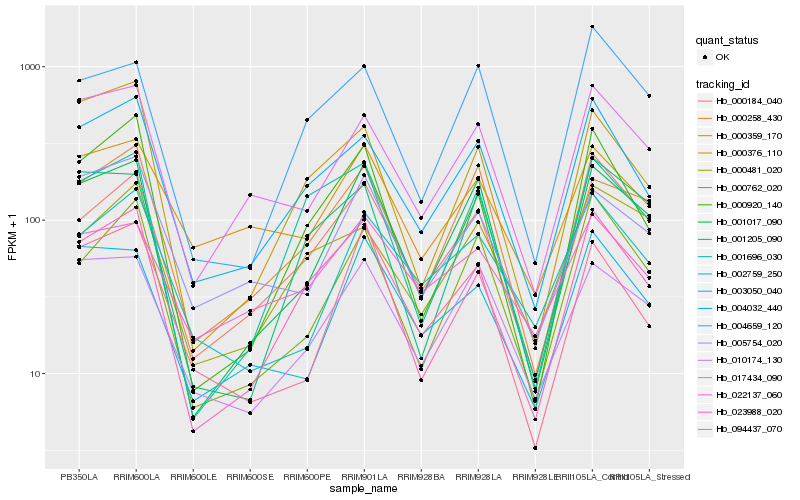
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_440 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 2 |
Hb_017434_090 |
0.0990466542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_023988_020 |
0.1018925394 |
- |
- |
PREDICTED: transmembrane protein 19 [Prunus mume] |
| 4 |
Hb_000184_040 |
0.1113812734 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 5 |
Hb_000376_110 |
0.1123940334 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Brachypodium distachyon] |
| 6 |
Hb_000920_140 |
0.118437187 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_000762_020 |
0.1227865878 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 8 |
Hb_001696_030 |
0.1256223682 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_010174_130 |
0.1270853447 |
- |
- |
PREDICTED: regulator of G-protein signaling 1 [Jatropha curcas] |
| 10 |
Hb_004659_120 |
0.1288858514 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000481_020 |
0.1328153259 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001017_090 |
0.1351442032 |
- |
- |
- |
| 13 |
Hb_002759_250 |
0.1356384473 |
rubber biosynthesis |
Gene Name: Phosphomevalonate kinase |
phosphomevalonate kinase [Hevea brasiliensis] |
| 14 |
Hb_005754_020 |
0.1372290388 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF4 [Jatropha curcas] |
| 15 |
Hb_000258_430 |
0.1372949686 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 16 |
Hb_000359_170 |
0.1389284708 |
- |
- |
hypothetical protein CISIN_1g014750mg [Citrus sinensis] |
| 17 |
Hb_001205_090 |
0.1410580423 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 18 |
Hb_003050_040 |
0.1436850304 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 19 |
Hb_022137_060 |
0.1436977795 |
- |
- |
1-acylglycerophosphocholine O-acyltransferase [Vernicia fordii] |
| 20 |
Hb_094437_070 |
0.1447951391 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |