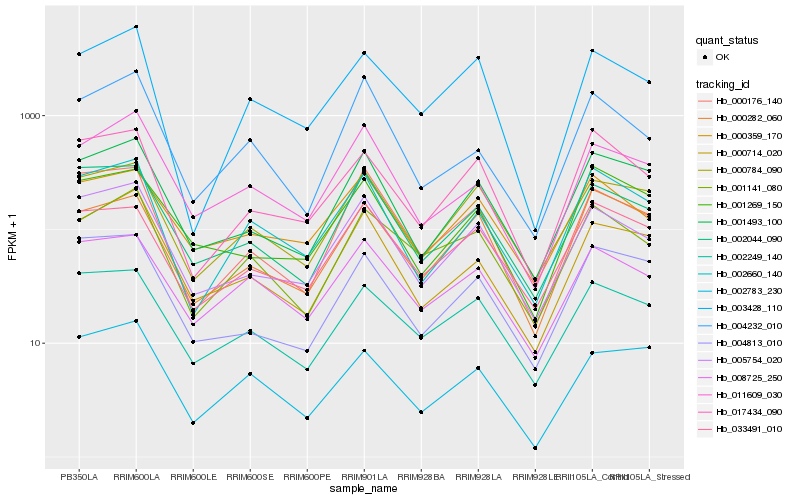
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002660_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s13060g [Populus trichocarpa] |
| 2 |
Hb_017434_090 |
0.090117945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001493_100 |
0.0917183544 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 34-like [Jatropha curcas] |
| 4 |
Hb_005754_020 |
0.1030448134 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF4 [Jatropha curcas] |
| 5 |
Hb_000714_020 |
0.1045791638 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 6 |
Hb_000784_090 |
0.1054751323 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_008725_250 |
0.1056978837 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_002044_090 |
0.1086848203 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004232_010 |
0.1088009415 |
- |
- |
PREDICTED: uncharacterized protein LOC105650678 [Jatropha curcas] |
| 10 |
Hb_033491_010 |
0.1088162306 |
- |
- |
Phosphatidylinositol N-acetylglucosaminyltransferase subunit C, putative [Ricinus communis] |
| 11 |
Hb_011609_030 |
0.1093068529 |
- |
- |
conserved hypothetical protein 13 [Hevea brasiliensis] |
| 12 |
Hb_000176_140 |
0.1130027027 |
- |
- |
PREDICTED: bystin [Jatropha curcas] |
| 13 |
Hb_000282_060 |
0.1133242541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638155 [Jatropha curcas] |
| 14 |
Hb_004813_010 |
0.1146504874 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_002249_140 |
0.1164089289 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 16 |
Hb_003428_110 |
0.1169943391 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 17 |
Hb_000359_170 |
0.117647823 |
- |
- |
hypothetical protein CISIN_1g014750mg [Citrus sinensis] |
| 18 |
Hb_001269_150 |
0.1186339978 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 19 |
Hb_002783_230 |
0.1188540915 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 20 |
Hb_001141_080 |
0.1208715726 |
- |
- |
PREDICTED: protein RDM1 [Jatropha curcas] |