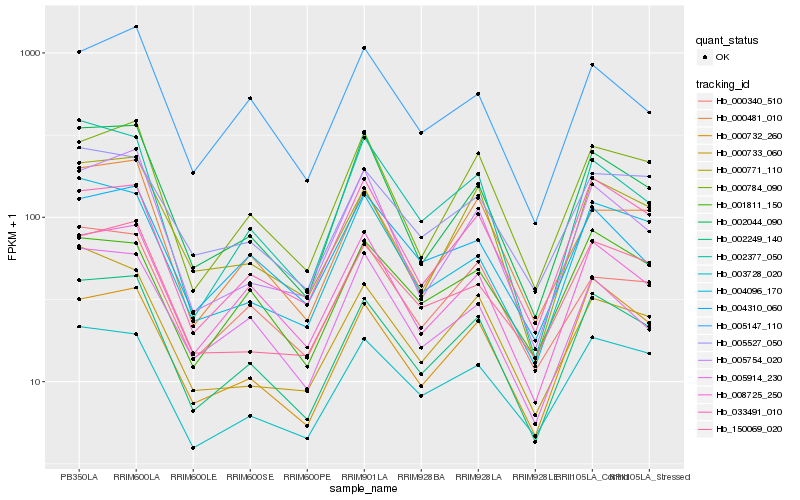
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 2 |
Hb_000771_110 |
0.0673334314 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 3 |
Hb_008725_250 |
0.0693068947 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_000732_260 |
0.0698416228 |
- |
- |
PREDICTED: telomerase Cajal body protein 1 [Jatropha curcas] |
| 5 |
Hb_002044_090 |
0.0713165686 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000481_010 |
0.0724731126 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP6 [Jatropha curcas] |
| 7 |
Hb_000784_090 |
0.0739436782 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_005527_050 |
0.0763365664 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 9 |
Hb_033491_010 |
0.0784967977 |
- |
- |
Phosphatidylinositol N-acetylglucosaminyltransferase subunit C, putative [Ricinus communis] |
| 10 |
Hb_003728_020 |
0.0787269389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_150069_020 |
0.0831306083 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000340_510 |
0.0836443262 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 13 |
Hb_005754_020 |
0.0841310295 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF4 [Jatropha curcas] |
| 14 |
Hb_004096_170 |
0.0843295004 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0275933 [Jatropha curcas] |
| 15 |
Hb_005147_110 |
0.0843368955 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 16 |
Hb_002377_050 |
0.0871773808 |
- |
- |
Agmatine deiminase, putative [Ricinus communis] |
| 17 |
Hb_001811_150 |
0.0872574289 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_005914_230 |
0.0896184689 |
- |
- |
PREDICTED: pre-mRNA-processing factor 19 homolog 1 [Jatropha curcas] |
| 19 |
Hb_000733_060 |
0.0902812669 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004310_060 |
0.0910761038 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |