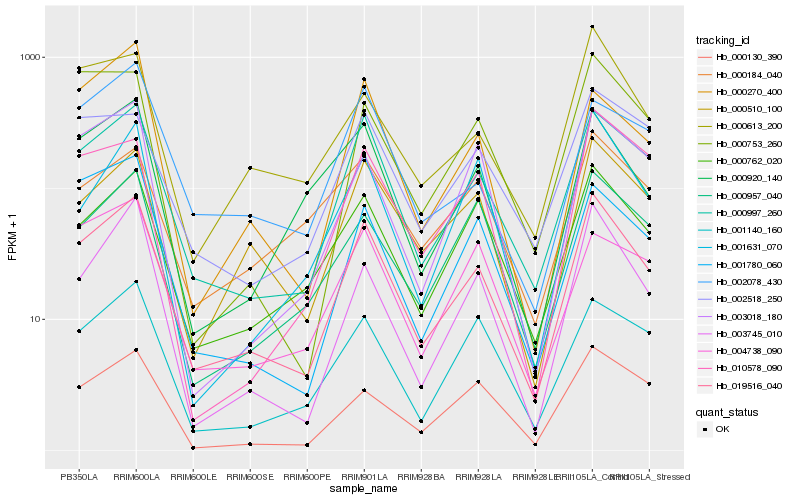
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019516_040 |
0.0 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 2 |
Hb_000997_260 |
0.0807854889 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000510_100 |
0.118207422 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000762_020 |
0.1207200161 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 5 |
Hb_003745_010 |
0.1329308766 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 6 |
Hb_001140_160 |
0.1363633517 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_000920_140 |
0.1370822261 |
- |
- |
annexin [Manihot esculenta] |
| 8 |
Hb_000130_390 |
0.1376283016 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 9 |
Hb_000613_200 |
0.1386845441 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 10 |
Hb_000957_040 |
0.1423928926 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 11 |
Hb_003018_180 |
0.1466915137 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 12 |
Hb_001780_060 |
0.1484415749 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 13 |
Hb_002518_250 |
0.1491427562 |
- |
- |
conserved hypothetical protein 10 [Hevea brasiliensis] |
| 14 |
Hb_001631_070 |
0.150573019 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_002078_430 |
0.1517346947 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 16 |
Hb_000270_400 |
0.1525090485 |
- |
- |
lipase, putative [Ricinus communis] |
| 17 |
Hb_000184_040 |
0.1549921215 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 18 |
Hb_010578_090 |
0.1553533684 |
- |
- |
HEV2.1 [Hevea brasiliensis] |
| 19 |
Hb_000753_260 |
0.1555032374 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 20 |
Hb_004738_090 |
0.1574732022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |