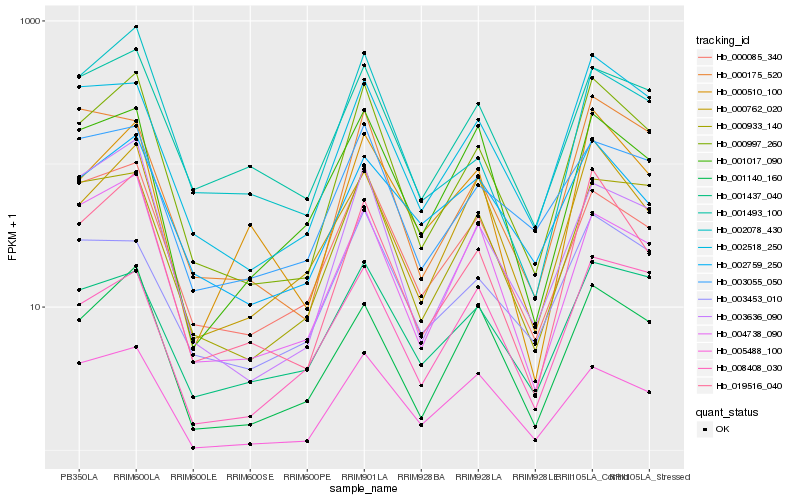
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_260 |
0.0 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_019516_040 |
0.0807854889 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 3 |
Hb_003636_090 |
0.1131012076 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3-like [Jatropha curcas] |
| 4 |
Hb_002518_250 |
0.1187496084 |
- |
- |
conserved hypothetical protein 10 [Hevea brasiliensis] |
| 5 |
Hb_001140_160 |
0.1222996895 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 6 |
Hb_000933_140 |
0.1250066921 |
- |
- |
PREDICTED: uncharacterized protein LOC105630501 [Jatropha curcas] |
| 7 |
Hb_000085_340 |
0.1272681876 |
- |
- |
PREDICTED: uncharacterized protein LOC105633038 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000762_020 |
0.1296225531 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 9 |
Hb_002078_430 |
0.1324354257 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 10 |
Hb_001493_100 |
0.1329617117 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 34-like [Jatropha curcas] |
| 11 |
Hb_000175_520 |
0.1363032545 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_004738_090 |
0.1378664959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001437_040 |
0.1384640166 |
- |
- |
PREDICTED: ATPase GET3-like [Vitis vinifera] |
| 14 |
Hb_000510_100 |
0.1389449223 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_005488_100 |
0.1397323976 |
- |
- |
hypothetical protein JCGZ_00295 [Jatropha curcas] |
| 16 |
Hb_003055_050 |
0.1429045796 |
- |
- |
hypothetical protein POPTR_0011s04510g [Populus trichocarpa] |
| 17 |
Hb_003453_010 |
0.1455995422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002759_250 |
0.1480842612 |
rubber biosynthesis |
Gene Name: Phosphomevalonate kinase |
phosphomevalonate kinase [Hevea brasiliensis] |
| 19 |
Hb_001017_090 |
0.1484540939 |
- |
- |
- |
| 20 |
Hb_008408_030 |
0.148496451 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |