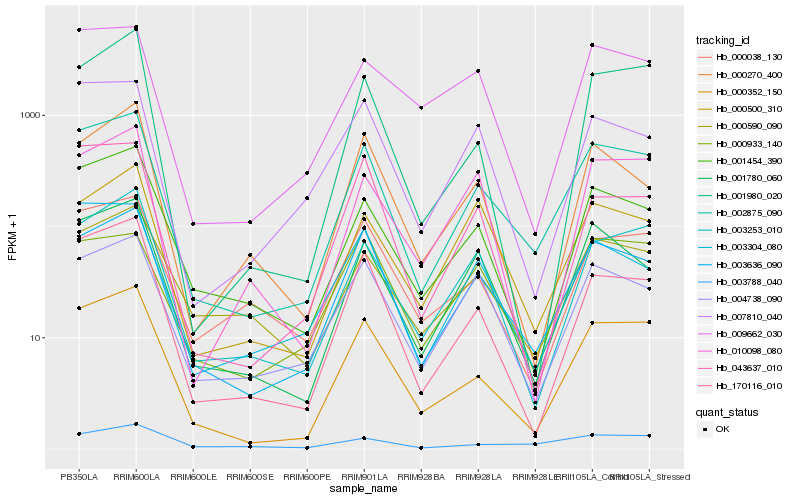
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002875_090 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003636_090 |
0.0755218176 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3-like [Jatropha curcas] |
| 3 |
Hb_000352_150 |
0.0850866158 |
- |
- |
hypothetical protein JCGZ_11641 [Jatropha curcas] |
| 4 |
Hb_001780_060 |
0.1125069938 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 5 |
Hb_001454_390 |
0.1141681227 |
- |
- |
8-kd dynein light chain, DLC8, pin [Thalassiosira pseudonana CCMP1335] |
| 6 |
Hb_170116_010 |
0.1167241724 |
- |
- |
Geranyl diphosphate synthase 1 isoform 2 [Theobroma cacao] |
| 7 |
Hb_003304_080 |
0.1252995944 |
transcription factor |
TF Family: bHLH |
lMYC3 [Hevea brasiliensis] |
| 8 |
Hb_003253_010 |
0.1285364245 |
- |
- |
PREDICTED: uncharacterized protein LOC105111108 [Populus euphratica] |
| 9 |
Hb_004738_090 |
0.1286663105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000038_130 |
0.1341612773 |
- |
- |
- |
| 11 |
Hb_007810_040 |
0.1367323544 |
- |
- |
PREDICTED: dolichyl-phosphate beta-glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_000590_090 |
0.1387167865 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_043637_010 |
0.1390571319 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0015s05220g, partial [Populus trichocarpa] |
| 14 |
Hb_001980_020 |
0.1427793115 |
- |
- |
- |
| 15 |
Hb_003788_040 |
0.1432831634 |
- |
- |
- |
| 16 |
Hb_009662_030 |
0.1453984427 |
- |
- |
hypothetical protein JCGZ_15748 [Jatropha curcas] |
| 17 |
Hb_010098_080 |
0.1457565208 |
- |
- |
PREDICTED: non-specific phospholipase C4-like [Jatropha curcas] |
| 18 |
Hb_000270_400 |
0.1472167798 |
- |
- |
lipase, putative [Ricinus communis] |
| 19 |
Hb_000500_310 |
0.1472617274 |
- |
- |
- |
| 20 |
Hb_000933_140 |
0.1479365491 |
- |
- |
PREDICTED: uncharacterized protein LOC105630501 [Jatropha curcas] |