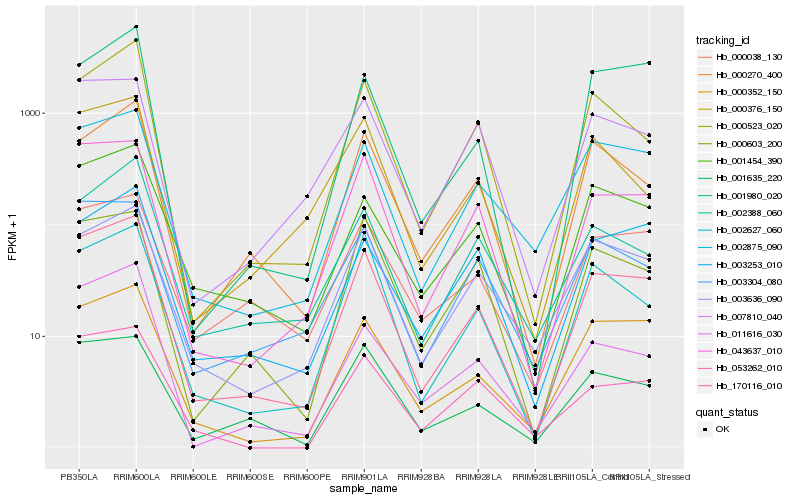
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170116_010 |
0.0 |
- |
- |
Geranyl diphosphate synthase 1 isoform 2 [Theobroma cacao] |
| 2 |
Hb_043637_010 |
0.0923891056 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0015s05220g, partial [Populus trichocarpa] |
| 3 |
Hb_000352_150 |
0.1046936729 |
- |
- |
hypothetical protein JCGZ_11641 [Jatropha curcas] |
| 4 |
Hb_000523_020 |
0.1090809649 |
- |
- |
PREDICTED: uncharacterized protein LOC105650610 [Jatropha curcas] |
| 5 |
Hb_001454_390 |
0.1103915527 |
- |
- |
8-kd dynein light chain, DLC8, pin [Thalassiosira pseudonana CCMP1335] |
| 6 |
Hb_002875_090 |
0.1167241724 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001635_220 |
0.1170518343 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 8 |
Hb_000270_400 |
0.1197492208 |
- |
- |
lipase, putative [Ricinus communis] |
| 9 |
Hb_053262_010 |
0.1233222828 |
- |
- |
- |
| 10 |
Hb_002627_060 |
0.1234951869 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 11 |
Hb_003636_090 |
0.1252759728 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3-like [Jatropha curcas] |
| 12 |
Hb_011616_030 |
0.1294345737 |
- |
- |
- |
| 13 |
Hb_003304_080 |
0.1297600184 |
transcription factor |
TF Family: bHLH |
lMYC3 [Hevea brasiliensis] |
| 14 |
Hb_000038_130 |
0.1320381438 |
- |
- |
- |
| 15 |
Hb_002388_060 |
0.1323219851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000603_200 |
0.1323942238 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 17 |
Hb_001980_020 |
0.1350377658 |
- |
- |
- |
| 18 |
Hb_000376_150 |
0.1364507935 |
- |
- |
O-acyltransferase WSD1 [Gossypium arboreum] |
| 19 |
Hb_007810_040 |
0.1403901381 |
- |
- |
PREDICTED: dolichyl-phosphate beta-glucosyltransferase [Jatropha curcas] |
| 20 |
Hb_003253_010 |
0.1407541324 |
- |
- |
PREDICTED: uncharacterized protein LOC105111108 [Populus euphratica] |