| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
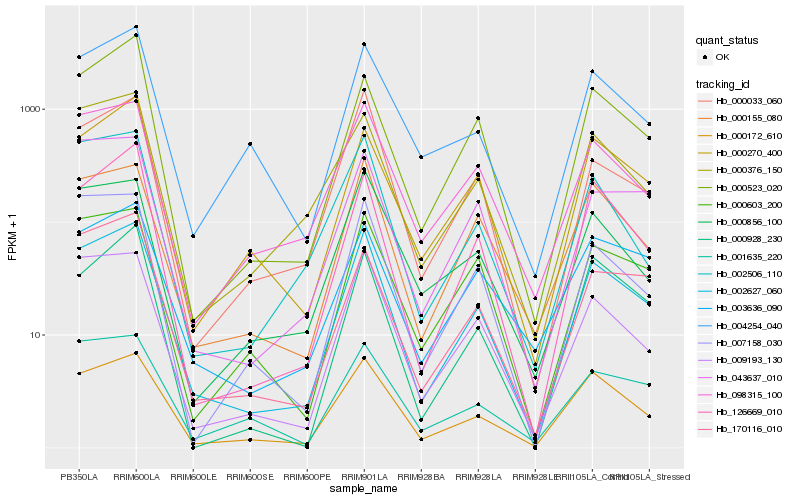
Hb_002627_060 |
0.0 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_000172_610 |
0.070733921 |
- |
- |
hypothetical protein POPTR_0017s02240g, partial [Populus trichocarpa] |
| 3 |
Hb_126669_010 |
0.1016617273 |
transcription factor |
TF Family: bHLH |
lMYC4 [Hevea brasiliensis] |
| 4 |
Hb_000155_080 |
0.1021040609 |
- |
- |
guanylate-binding family protein [Populus trichocarpa] |
| 5 |
Hb_009193_130 |
0.1032558253 |
- |
- |
hypothetical protein RCOM_1620550 [Ricinus communis] |
| 6 |
Hb_098315_100 |
0.1106783002 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000270_400 |
0.1123692709 |
- |
- |
lipase, putative [Ricinus communis] |
| 8 |
Hb_000033_060 |
0.1124852348 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 9 |
Hb_000376_150 |
0.1143025506 |
- |
- |
O-acyltransferase WSD1 [Gossypium arboreum] |
| 10 |
Hb_000603_200 |
0.1152681197 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 11 |
Hb_000523_020 |
0.1159994765 |
- |
- |
PREDICTED: uncharacterized protein LOC105650610 [Jatropha curcas] |
| 12 |
Hb_002506_110 |
0.1216927777 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 13 |
Hb_000856_100 |
0.1229244936 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 14 |
Hb_170116_010 |
0.1234951869 |
- |
- |
Geranyl diphosphate synthase 1 isoform 2 [Theobroma cacao] |
| 15 |
Hb_007158_030 |
0.1235579487 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_003636_090 |
0.1241754457 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3-like [Jatropha curcas] |
| 17 |
Hb_004254_040 |
0.125821065 |
- |
- |
PREDICTED: uncharacterized protein LOC105650678 [Jatropha curcas] |
| 18 |
Hb_000928_230 |
0.1283440851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001635_220 |
0.1288069782 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 20 |
Hb_043637_010 |
0.1293807492 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0015s05220g, partial [Populus trichocarpa] |