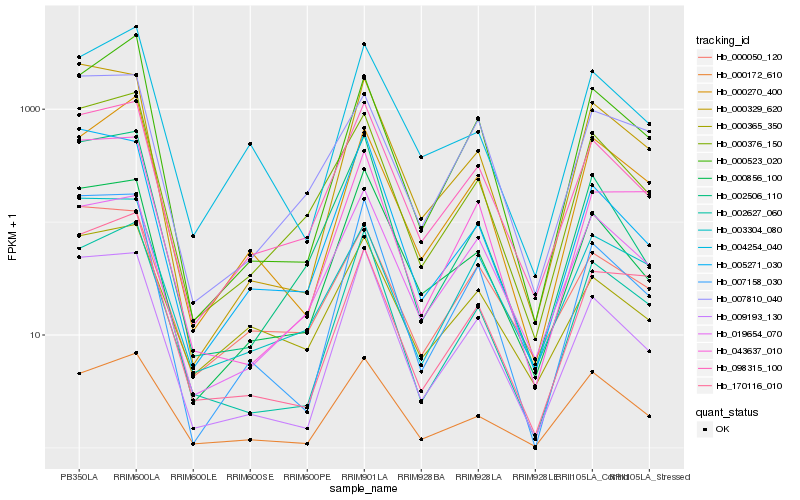
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000376_150 |
0.0 |
- |
- |
O-acyltransferase WSD1 [Gossypium arboreum] |
| 2 |
Hb_098315_100 |
0.0872884499 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002506_110 |
0.09059302 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 4 |
Hb_003304_080 |
0.1111155391 |
transcription factor |
TF Family: bHLH |
lMYC3 [Hevea brasiliensis] |
| 5 |
Hb_002627_060 |
0.1143025506 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 6 |
Hb_000172_610 |
0.1158553997 |
- |
- |
hypothetical protein POPTR_0017s02240g, partial [Populus trichocarpa] |
| 7 |
Hb_000050_120 |
0.1184464328 |
- |
- |
PREDICTED: uncharacterized protein LOC105630790 [Jatropha curcas] |
| 8 |
Hb_000365_350 |
0.1210691012 |
- |
- |
PREDICTED: uncharacterized protein LOC105649068 [Jatropha curcas] |
| 9 |
Hb_007810_040 |
0.1218301854 |
- |
- |
PREDICTED: dolichyl-phosphate beta-glucosyltransferase [Jatropha curcas] |
| 10 |
Hb_000523_020 |
0.1232018245 |
- |
- |
PREDICTED: uncharacterized protein LOC105650610 [Jatropha curcas] |
| 11 |
Hb_000856_100 |
0.1271403444 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 12 |
Hb_000270_400 |
0.1286789567 |
- |
- |
lipase, putative [Ricinus communis] |
| 13 |
Hb_019654_070 |
0.1303188236 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 14 |
Hb_007158_030 |
0.1312749129 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000329_620 |
0.1317688029 |
- |
- |
PREDICTED: uncharacterized protein LOC105643215 [Jatropha curcas] |
| 16 |
Hb_009193_130 |
0.1326110029 |
- |
- |
hypothetical protein RCOM_1620550 [Ricinus communis] |
| 17 |
Hb_004254_040 |
0.1333693082 |
- |
- |
PREDICTED: uncharacterized protein LOC105650678 [Jatropha curcas] |
| 18 |
Hb_170116_010 |
0.1364507935 |
- |
- |
Geranyl diphosphate synthase 1 isoform 2 [Theobroma cacao] |
| 19 |
Hb_043637_010 |
0.1373813906 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0015s05220g, partial [Populus trichocarpa] |
| 20 |
Hb_005271_030 |
0.1378955804 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |