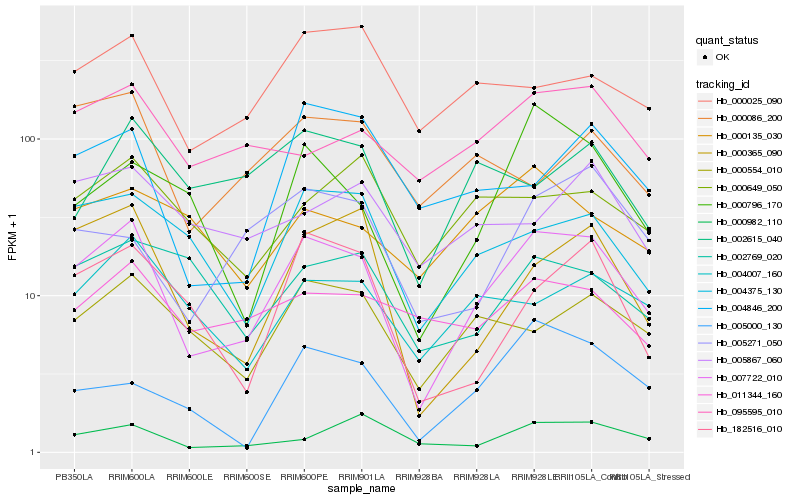
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007722_010 |
0.0 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 2 |
Hb_000365_090 |
0.1774595231 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_004375_130 |
0.1825212835 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000554_010 |
0.1852092817 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_004007_160 |
0.1928906639 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 6 |
Hb_182516_010 |
0.193687831 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 7 |
Hb_095595_010 |
0.2012617567 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 8 |
Hb_000025_090 |
0.2015359328 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 9 |
Hb_000796_170 |
0.2031761687 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 10 |
Hb_005000_130 |
0.2051754687 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 11 |
Hb_011344_160 |
0.2055098293 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 12 |
Hb_004846_200 |
0.2056854741 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 13 |
Hb_000649_050 |
0.2094568836 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000982_110 |
0.2095334534 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_005271_050 |
0.2123975631 |
- |
- |
Tubulin beta-8 chain -like protein [Gossypium arboreum] |
| 16 |
Hb_000135_030 |
0.2128872863 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002769_020 |
0.2143135821 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 18 |
Hb_000086_200 |
0.2172991602 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_002615_040 |
0.2184844026 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 20 |
Hb_005867_060 |
0.2202845016 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase [Jatropha curcas] |