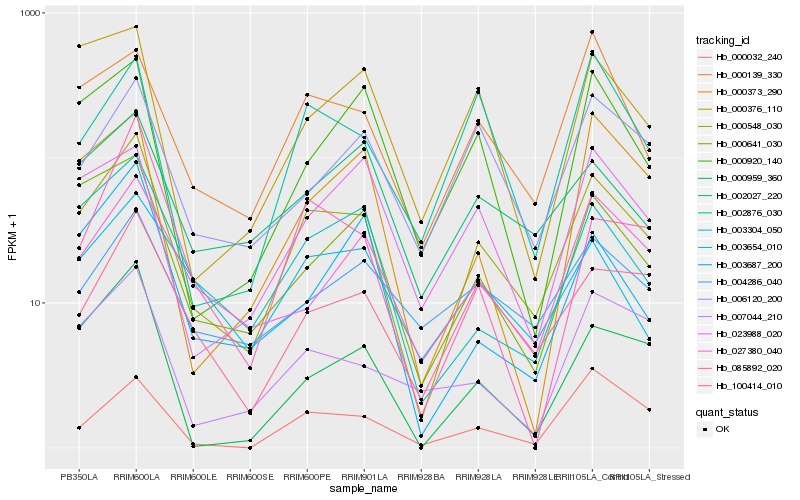
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000548_030 |
0.0 |
rubber biosynthesis |
Gene Name: CPT5 |
PREDICTED: dehydrodolichyl diphosphate synthase 6-like [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_003304_050 |
0.1587047628 |
- |
- |
PREDICTED: uncharacterized protein LOC105633679 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000641_030 |
0.1612080186 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 4 |
Hb_100414_010 |
0.1667230929 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_000139_330 |
0.1672986505 |
- |
- |
short chain dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000959_360 |
0.1732237942 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_003654_010 |
0.1758918854 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 8 |
Hb_027380_040 |
0.1767997346 |
- |
- |
hypothetical protein B456_011G046500, partial [Gossypium raimondii] |
| 9 |
Hb_002027_220 |
0.1803234153 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_007044_210 |
0.1847413329 |
transcription factor |
TF Family: MYB |
PREDICTED: protein rough sheath 2 homolog [Populus euphratica] |
| 11 |
Hb_004286_040 |
0.1847520433 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006120_200 |
0.1869412348 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 13 |
Hb_000920_140 |
0.1890857091 |
- |
- |
annexin [Manihot esculenta] |
| 14 |
Hb_000376_110 |
0.1895504446 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Brachypodium distachyon] |
| 15 |
Hb_023988_020 |
0.1913738132 |
- |
- |
PREDICTED: transmembrane protein 19 [Prunus mume] |
| 16 |
Hb_000373_290 |
0.1928700115 |
- |
- |
RecName: Full=Citrate-binding protein; Flags: Precursor [Hevea brasiliensis] |
| 17 |
Hb_000032_240 |
0.1956936076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002876_030 |
0.1960052779 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 19 |
Hb_085892_020 |
0.197655793 |
- |
- |
- |
| 20 |
Hb_003687_200 |
0.1997983143 |
- |
- |
PREDICTED: uncharacterized protein LOC105632139 [Jatropha curcas] |