| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
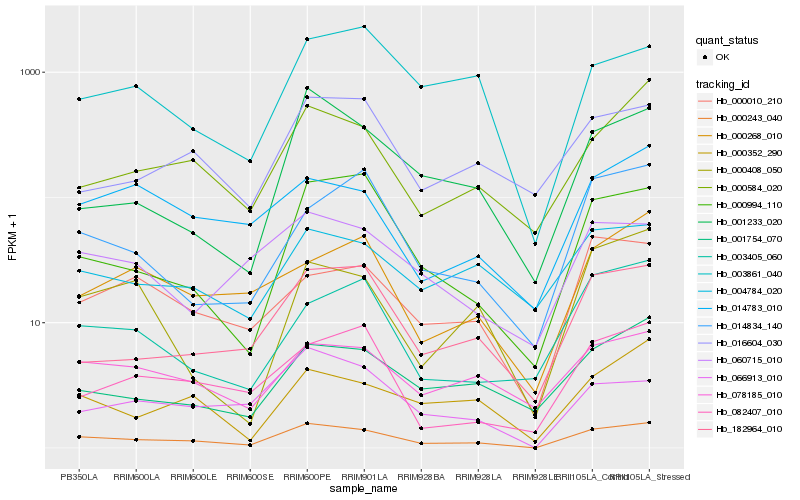
Hb_000243_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000994_110 |
0.1537284414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_078185_010 |
0.1588855535 |
- |
- |
- |
| 4 |
Hb_000584_020 |
0.1594079544 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 5 |
Hb_000352_290 |
0.1631027348 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 6 |
Hb_182964_010 |
0.1650327829 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000010_210 |
0.1660015542 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 8 |
Hb_066913_010 |
0.1671657761 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 9 |
Hb_001754_070 |
0.1689581568 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 10 |
Hb_014783_010 |
0.1735392392 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_082407_010 |
0.1750118941 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 12 |
Hb_001233_020 |
0.175617148 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 13 |
Hb_004784_020 |
0.1758807711 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 14 |
Hb_060715_010 |
0.1775984702 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 15 |
Hb_014834_140 |
0.1776083005 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 16 |
Hb_000268_010 |
0.1790010974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003405_060 |
0.1803807889 |
- |
- |
- |
| 18 |
Hb_003861_040 |
0.1822180605 |
- |
- |
hypothetical protein JCGZ_16325 [Jatropha curcas] |
| 19 |
Hb_016604_030 |
0.1829713538 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_000408_050 |
0.1843499792 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |