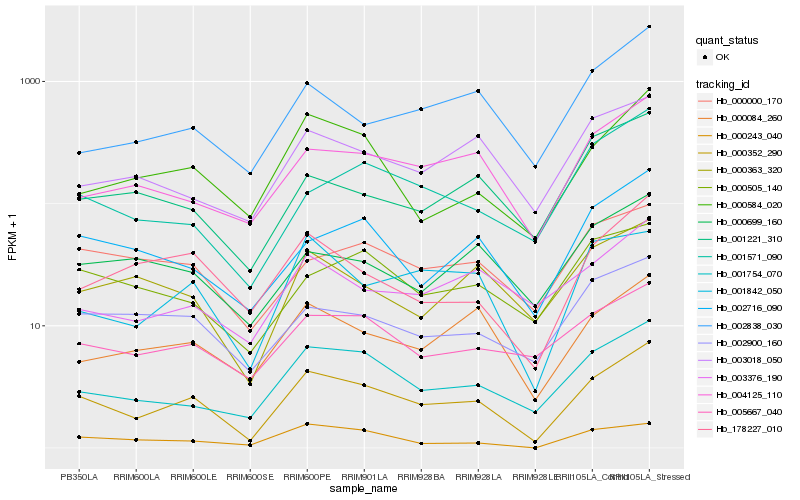
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_290 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 2 |
Hb_001754_070 |
0.1355227004 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 3 |
Hb_001842_050 |
0.1462915588 |
- |
- |
PREDICTED: protein BUD31 homolog 1-like [Jatropha curcas] |
| 4 |
Hb_004125_110 |
0.1508549672 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 5 |
Hb_000584_020 |
0.1512544218 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 6 |
Hb_000084_260 |
0.1519010252 |
- |
- |
PREDICTED: replication protein A 14 kDa subunit A-like [Jatropha curcas] |
| 7 |
Hb_178227_010 |
0.1558500902 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 8 |
Hb_002716_090 |
0.1571231908 |
- |
- |
U6 snRNA-associated Sm-like protein LSm8 [Hevea brasiliensis] |
| 9 |
Hb_002900_160 |
0.1614431556 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 10 |
Hb_005667_040 |
0.1616743988 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Jatropha curcas] |
| 11 |
Hb_000243_040 |
0.1631027348 |
- |
- |
- |
| 12 |
Hb_000363_320 |
0.1645140778 |
- |
- |
PREDICTED: V-type proton ATPase subunit e1 [Jatropha curcas] |
| 13 |
Hb_001571_090 |
0.1681794636 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 14 |
Hb_003018_050 |
0.1694259831 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 15 |
Hb_002838_030 |
0.1698269151 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 16 |
Hb_003376_190 |
0.1699594577 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_001221_310 |
0.1712590447 |
- |
- |
sec61 gamma subunit, putative [Ricinus communis] |
| 18 |
Hb_000000_170 |
0.1714138545 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 19 |
Hb_000699_160 |
0.1715374222 |
- |
- |
PREDICTED: C-Myc-binding protein homolog [Jatropha curcas] |
| 20 |
Hb_000505_140 |
0.1734875353 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 3 [Populus euphratica] |