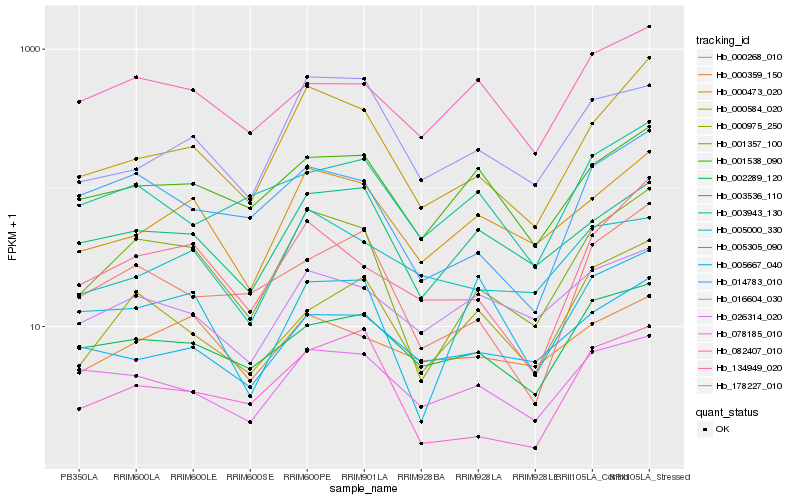
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_100 |
0.0 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 2 |
Hb_000584_020 |
0.1160809035 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 3 |
Hb_014783_010 |
0.1341875438 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000473_020 |
0.1424411757 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 5 |
Hb_178227_010 |
0.1465756975 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 6 |
Hb_003536_110 |
0.1466492802 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 7 |
Hb_000268_010 |
0.1559205898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001538_090 |
0.1595601134 |
- |
- |
40S ribosomal protein S29 [Morus notabilis] |
| 9 |
Hb_082407_010 |
0.1605240025 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 10 |
Hb_078185_010 |
0.1632898528 |
- |
- |
- |
| 11 |
Hb_002289_120 |
0.1634703435 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005305_090 |
0.1658541847 |
- |
- |
PREDICTED: uncharacterized protein LOC105112272 [Populus euphratica] |
| 13 |
Hb_000975_250 |
0.1662831401 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 4, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_026314_020 |
0.1700207476 |
- |
- |
hypothetical protein POPTR_0019s10050g [Populus trichocarpa] |
| 15 |
Hb_016604_030 |
0.1723262683 |
- |
- |
histone H4 [Zea mays] |
| 16 |
Hb_134949_020 |
0.1743560886 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 [Cucumis sativus] |
| 17 |
Hb_005667_040 |
0.1755764515 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Jatropha curcas] |
| 18 |
Hb_000359_150 |
0.1765039122 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005000_330 |
0.1777904072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003943_130 |
0.1783849513 |
- |
- |
PREDICTED: uncharacterized protein LOC105636341 [Jatropha curcas] |