| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
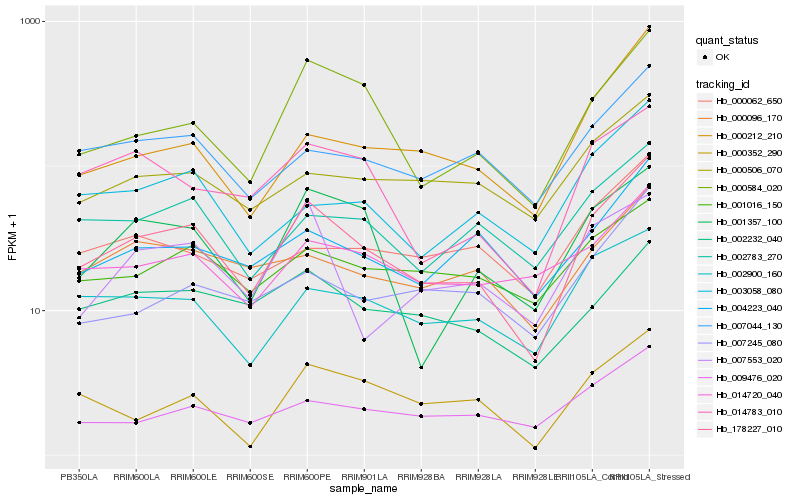
Hb_178227_010 |
0.0 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 2 |
Hb_000584_020 |
0.122813309 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 3 |
Hb_002900_160 |
0.1387304085 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 4 |
Hb_014783_010 |
0.1408608042 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_007044_130 |
0.1420719275 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 6 |
Hb_002783_270 |
0.1436322971 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 7 |
Hb_009476_020 |
0.1449833591 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 8 |
Hb_001357_100 |
0.1465756975 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 9 |
Hb_000096_170 |
0.1477704573 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 10 |
Hb_003058_080 |
0.1489450861 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 11 |
Hb_004223_040 |
0.1495397728 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 12 |
Hb_002232_040 |
0.1511942094 |
- |
- |
- |
| 13 |
Hb_014720_040 |
0.1524117528 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001016_150 |
0.1535784517 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 15 |
Hb_000506_070 |
0.1540360697 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 16 |
Hb_000352_290 |
0.1558500902 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 17 |
Hb_007245_080 |
0.1580565303 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 18 |
Hb_000212_210 |
0.1592915441 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000062_650 |
0.1593240923 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 20 |
Hb_007553_020 |
0.1605885191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |