| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
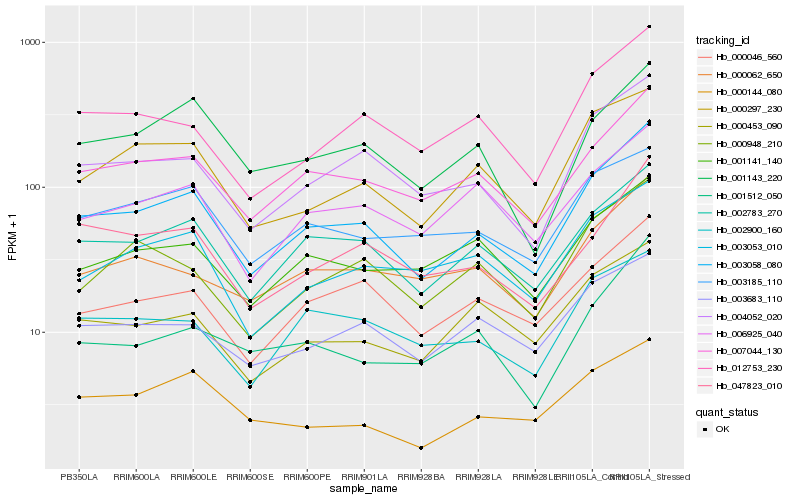
Hb_003058_080 |
0.0 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 2 |
Hb_002783_270 |
0.0790267181 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 3 |
Hb_007044_130 |
0.0828824852 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 4 |
Hb_004052_020 |
0.0834820718 |
- |
- |
Mitochondrial import receptor subunit TOM5 [Theobroma cacao] |
| 5 |
Hb_012753_230 |
0.1010473452 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 6 |
Hb_000046_560 |
0.1014719008 |
- |
- |
PREDICTED: uncharacterized protein LOC105118519 [Populus euphratica] |
| 7 |
Hb_047823_010 |
0.1034746575 |
- |
- |
hypothetical protein CARUB_v10024013mg [Capsella rubella] |
| 8 |
Hb_000297_230 |
0.1043791226 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 9 |
Hb_000062_650 |
0.1082005468 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 10 |
Hb_000948_210 |
0.1083723502 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_006925_040 |
0.1123604557 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 12 |
Hb_002900_160 |
0.1126392514 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 13 |
Hb_001512_050 |
0.1128472527 |
- |
- |
copine, putative [Ricinus communis] |
| 14 |
Hb_001143_220 |
0.1130297476 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 15 |
Hb_003683_110 |
0.1140884118 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 16 |
Hb_000453_090 |
0.1143565186 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 17 |
Hb_003053_010 |
0.1156331176 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000144_080 |
0.1161245166 |
- |
- |
PREDICTED: uncharacterized protein LOC104609774 [Nelumbo nucifera] |
| 19 |
Hb_003185_110 |
0.116157492 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 7-2 [Nelumbo nucifera] |
| 20 |
Hb_001141_140 |
0.1194004066 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |