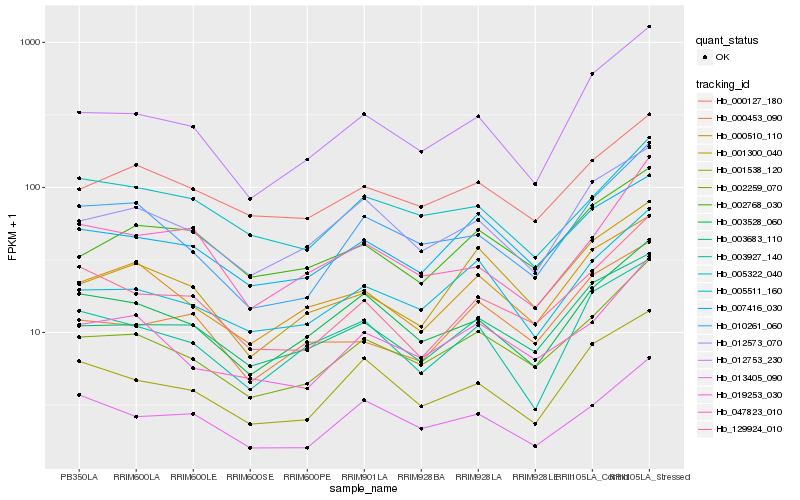
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129924_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630505 [Jatropha curcas] |
| 2 |
Hb_002259_070 |
0.0934596266 |
- |
- |
taz protein, putative [Ricinus communis] |
| 3 |
Hb_013405_090 |
0.0990633464 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001538_120 |
0.1021341581 |
- |
- |
PREDICTED: FAD-linked sulfhydryl oxidase ERV1 [Jatropha curcas] |
| 5 |
Hb_003528_060 |
0.1042911366 |
- |
- |
PREDICTED: uncharacterized protein LOC105638256 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003683_110 |
0.1057165456 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 7 |
Hb_012753_230 |
0.106544834 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 8 |
Hb_047823_010 |
0.1077705442 |
- |
- |
hypothetical protein CARUB_v10024013mg [Capsella rubella] |
| 9 |
Hb_007416_030 |
0.1109736338 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 10 [Jatropha curcas] |
| 10 |
Hb_010261_060 |
0.1110941723 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_019253_030 |
0.1114585177 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-3-like [Citrus sinensis] |
| 12 |
Hb_000453_090 |
0.1165649402 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 13 |
Hb_000510_110 |
0.1166716148 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 14 |
Hb_003927_140 |
0.1213932407 |
- |
- |
PREDICTED: nudix hydrolase 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000127_180 |
0.1237448843 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 16 |
Hb_005322_040 |
0.1240562425 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 17 |
Hb_001300_040 |
0.1247244932 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 18 |
Hb_002768_030 |
0.1254466693 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 19 |
Hb_012573_070 |
0.1254908337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005511_160 |
0.1261552924 |
- |
- |
OB-fold nucleic acid binding domain-containing protein [Theobroma cacao] |