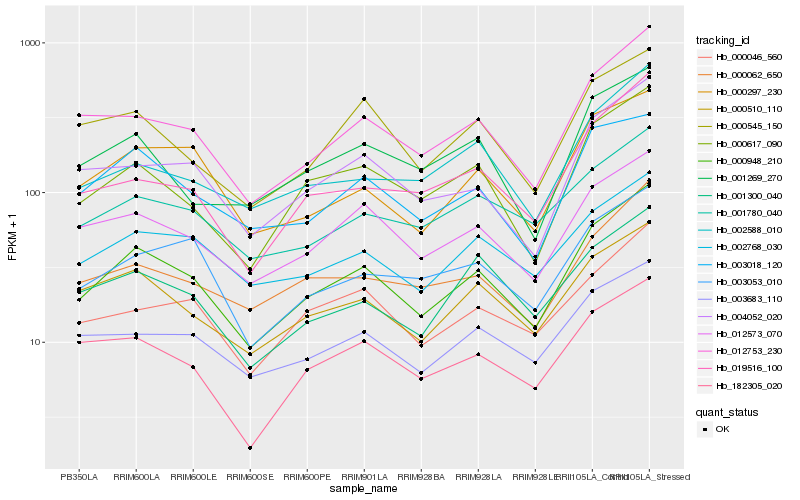
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000948_210 |
0.0 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000617_090 |
0.0781332635 |
- |
- |
hypoia-responsive family protein 4 [Hevea brasiliensis] |
| 3 |
Hb_012753_230 |
0.079482676 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 4 |
Hb_012573_070 |
0.0856229767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004052_020 |
0.0875406967 |
- |
- |
Mitochondrial import receptor subunit TOM5 [Theobroma cacao] |
| 6 |
Hb_000510_110 |
0.0893969273 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 7 |
Hb_000297_230 |
0.0906034474 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 8 |
Hb_000062_650 |
0.0937465669 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 9 |
Hb_002768_030 |
0.0941505805 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 10 |
Hb_003018_120 |
0.0946092139 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 11 |
Hb_000046_560 |
0.0949402132 |
- |
- |
PREDICTED: uncharacterized protein LOC105118519 [Populus euphratica] |
| 12 |
Hb_003683_110 |
0.0952747127 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 13 |
Hb_019516_100 |
0.0960948704 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 14 |
Hb_001780_040 |
0.0967913218 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 15 |
Hb_182305_020 |
0.0970908577 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000545_150 |
0.0976309485 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 17 |
Hb_003053_010 |
0.0985983983 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001300_040 |
0.0998932992 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 19 |
Hb_001269_270 |
0.1002121309 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 20 |
Hb_002588_010 |
0.1002514793 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |