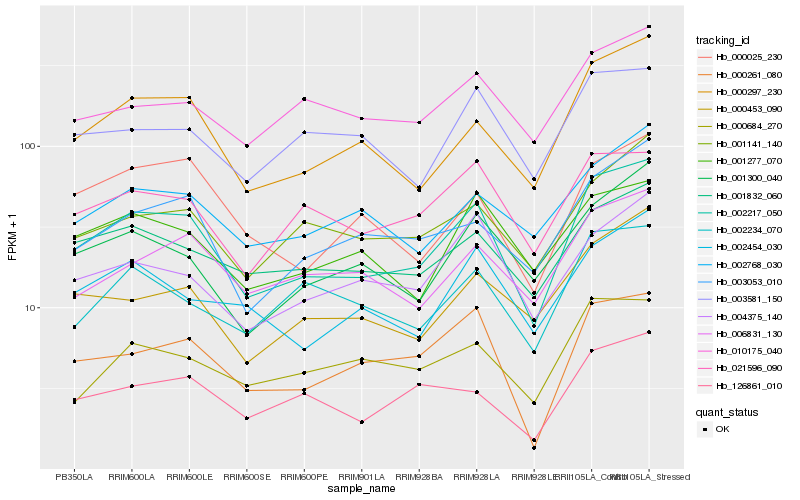
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104607091 isoform X1 [Nelumbo nucifera] |
| 2 |
Hb_000297_230 |
0.0930518939 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 3 |
Hb_000261_080 |
0.1061503855 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003053_010 |
0.1118748032 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001832_060 |
0.1139673399 |
- |
- |
PREDICTED: translation machinery-associated protein 22 [Jatropha curcas] |
| 6 |
Hb_001300_040 |
0.1151668986 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 7 |
Hb_006831_130 |
0.1167150741 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_001141_140 |
0.1167876482 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |
| 9 |
Hb_002454_030 |
0.1171852454 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B [Jatropha curcas] |
| 10 |
Hb_000453_090 |
0.1182670155 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 11 |
Hb_021596_090 |
0.118703463 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8-B [Jatropha curcas] |
| 12 |
Hb_004375_140 |
0.1189481714 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_126861_010 |
0.1191552964 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 14 |
Hb_002768_030 |
0.1200993807 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 15 |
Hb_001277_070 |
0.1208529386 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002234_070 |
0.1226519057 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000025_230 |
0.1246378412 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |
| 18 |
Hb_010175_040 |
0.1247314305 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6a, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000684_270 |
0.1251960493 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003581_150 |
0.1257626483 |
- |
- |
hypothetical protein CICLE_v10032366mg [Citrus clementina] |