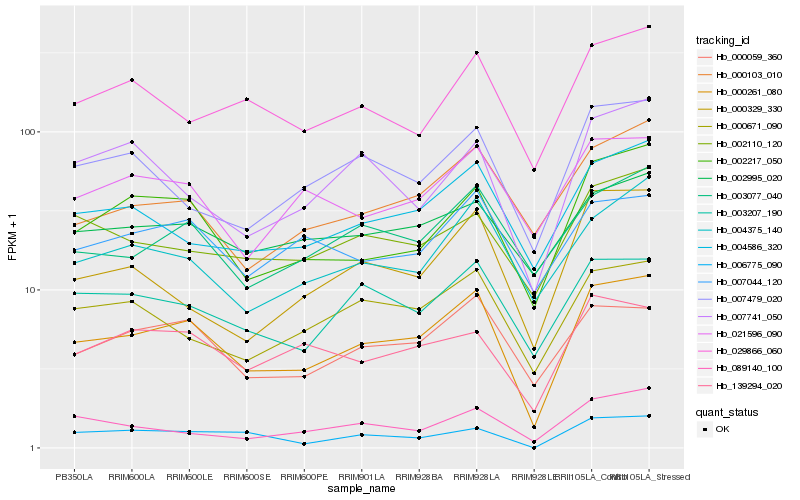
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002217_050 |
0.1061503855 |
- |
- |
PREDICTED: uncharacterized protein LOC104607091 isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_000059_360 |
0.1162040776 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 4 |
Hb_004586_320 |
0.1197739892 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 5 |
Hb_004375_140 |
0.1229246078 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003077_040 |
0.1247260595 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 7 |
Hb_000103_010 |
0.1261800637 |
- |
- |
20S proteasome beta subunit D2 [Hevea brasiliensis] |
| 8 |
Hb_003207_190 |
0.1270628389 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Populus euphratica] |
| 9 |
Hb_000671_090 |
0.128719238 |
- |
- |
PREDICTED: melanoma-associated antigen 1 [Jatropha curcas] |
| 10 |
Hb_007479_020 |
0.131366923 |
- |
- |
PREDICTED: CMP-sialic acid transporter 5 [Jatropha curcas] |
| 11 |
Hb_139294_020 |
0.1343794643 |
- |
- |
vat protein [Cucumis melo] |
| 12 |
Hb_021596_090 |
0.1353590533 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8-B [Jatropha curcas] |
| 13 |
Hb_000329_330 |
0.1363074445 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 14 |
Hb_029866_060 |
0.1363781784 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 15 |
Hb_089140_100 |
0.1369826621 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 16 |
Hb_007044_120 |
0.1375069212 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 17 |
Hb_002995_020 |
0.1375946729 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 18 |
Hb_006775_090 |
0.1378385684 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_002110_120 |
0.1380344587 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 20 |
Hb_007741_050 |
0.1384724258 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |