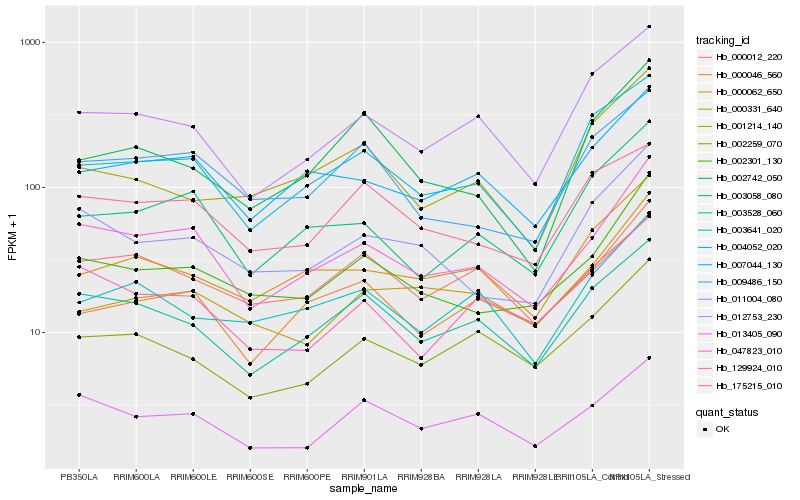
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X3 [Populus euphratica] |
| 2 |
Hb_011004_080 |
0.0849997977 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Cucumis sativus] |
| 3 |
Hb_047823_010 |
0.091409395 |
- |
- |
hypothetical protein CARUB_v10024013mg [Capsella rubella] |
| 4 |
Hb_009486_150 |
0.1085407248 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 5 |
Hb_000331_640 |
0.1147099298 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000062_650 |
0.1184046122 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 7 |
Hb_007044_130 |
0.1230730673 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 8 |
Hb_003528_060 |
0.1260012609 |
- |
- |
PREDICTED: uncharacterized protein LOC105638256 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003058_080 |
0.1272526008 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 10 |
Hb_001214_140 |
0.127743114 |
- |
- |
60S ribosomal protein L27a, putative [Ricinus communis] |
| 11 |
Hb_004052_020 |
0.1277855497 |
- |
- |
Mitochondrial import receptor subunit TOM5 [Theobroma cacao] |
| 12 |
Hb_003641_020 |
0.1280897277 |
- |
- |
hypothetical protein POPTR_0006s13960g [Populus trichocarpa] |
| 13 |
Hb_002742_050 |
0.1283139391 |
- |
- |
hypothetical protein CICLE_v10002863mg [Citrus clementina] |
| 14 |
Hb_000012_220 |
0.1298889111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002259_070 |
0.1305010952 |
- |
- |
taz protein, putative [Ricinus communis] |
| 16 |
Hb_129924_010 |
0.1337591024 |
- |
- |
PREDICTED: uncharacterized protein LOC105630505 [Jatropha curcas] |
| 17 |
Hb_175215_010 |
0.1340516224 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 18 |
Hb_000046_560 |
0.1342132058 |
- |
- |
PREDICTED: uncharacterized protein LOC105118519 [Populus euphratica] |
| 19 |
Hb_012753_230 |
0.134750698 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 20 |
Hb_013405_090 |
0.1363263589 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X1 [Jatropha curcas] |