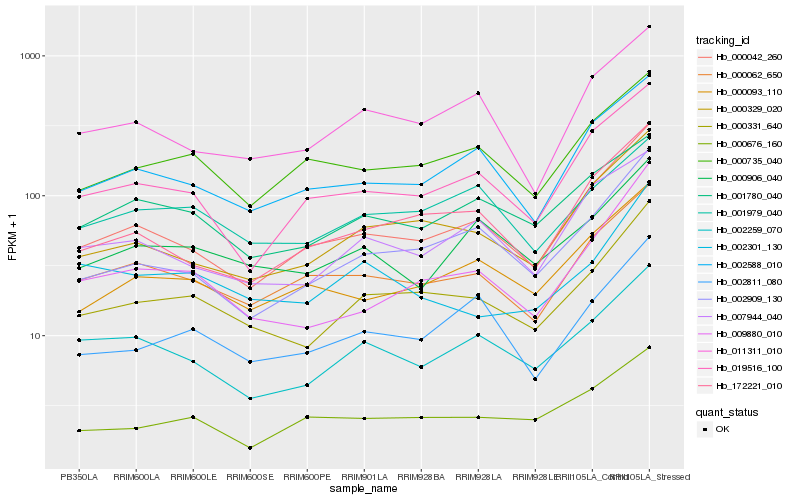
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_640 |
0.0 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000329_020 |
0.0861213289 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 3 |
Hb_002909_130 |
0.09415638 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_000042_260 |
0.0981077921 |
- |
- |
PREDICTED: uncharacterized protein LOC105632814 [Jatropha curcas] |
| 5 |
Hb_009880_010 |
0.0988609367 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 6 |
Hb_000062_650 |
0.1039662381 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 7 |
Hb_002811_080 |
0.1062906892 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 8 |
Hb_002588_010 |
0.1069939847 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 9 |
Hb_002259_070 |
0.1084229365 |
- |
- |
taz protein, putative [Ricinus communis] |
| 10 |
Hb_172221_010 |
0.10878345 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 11 |
Hb_000093_110 |
0.1091935537 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 12 |
Hb_000735_040 |
0.1102137338 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 13 |
Hb_011311_010 |
0.111025383 |
- |
- |
60S ribosomal protein L27C [Hevea brasiliensis] |
| 14 |
Hb_001979_040 |
0.1127448964 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1-like [Gossypium raimondii] |
| 15 |
Hb_002301_130 |
0.1147099298 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X3 [Populus euphratica] |
| 16 |
Hb_001780_040 |
0.1149751946 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 17 |
Hb_007944_040 |
0.1161218592 |
- |
- |
60S ribosomal protein L18aB [Hevea brasiliensis] |
| 18 |
Hb_000676_160 |
0.1161225116 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 19 |
Hb_019516_100 |
0.1171078656 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 20 |
Hb_000906_040 |
0.1178639584 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |