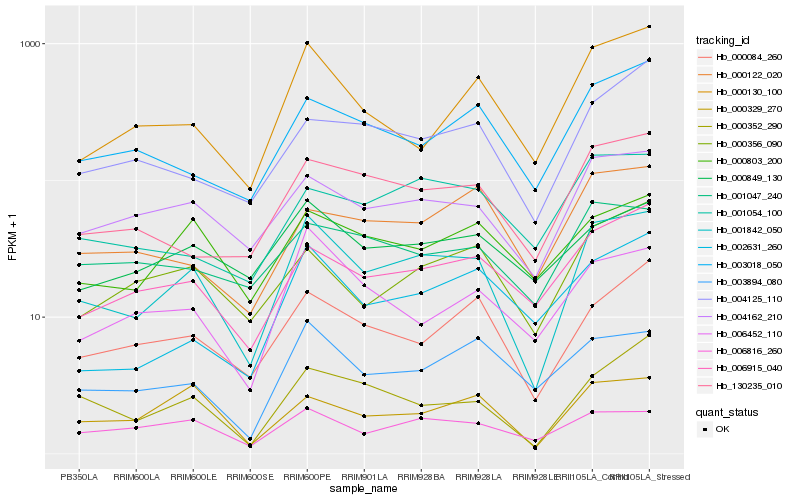
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001842_050 |
0.0 |
- |
- |
PREDICTED: protein BUD31 homolog 1-like [Jatropha curcas] |
| 2 |
Hb_004162_210 |
0.133258745 |
- |
- |
Cyclin-dependent kinases regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_000352_290 |
0.1462915588 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 4 |
Hb_000084_260 |
0.1469461609 |
- |
- |
PREDICTED: replication protein A 14 kDa subunit A-like [Jatropha curcas] |
| 5 |
Hb_006915_040 |
0.1480713945 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 6 |
Hb_130235_010 |
0.1498845196 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 7 |
Hb_000329_270 |
0.1522205894 |
- |
- |
PREDICTED: spindle and kinetochore-associated protein 1 homolog [Jatropha curcas] |
| 8 |
Hb_003894_080 |
0.1523726597 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Vitis vinifera] |
| 9 |
Hb_006452_110 |
0.1544934052 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 10 |
Hb_006816_260 |
0.1572405764 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 11 |
Hb_000130_100 |
0.1576236404 |
- |
- |
hypothetical protein CICLE_v10022662mg [Citrus clementina] |
| 12 |
Hb_003018_050 |
0.1583726022 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 13 |
Hb_000803_200 |
0.1594631292 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 14 |
Hb_000356_090 |
0.1601448711 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 15 |
Hb_000849_130 |
0.1622056478 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 16 |
Hb_002631_260 |
0.1664875371 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 17 |
Hb_001054_100 |
0.1674635544 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 18 |
Hb_001047_240 |
0.169840042 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 19 |
Hb_004125_110 |
0.1699716644 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 20 |
Hb_000122_020 |
0.170518957 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |