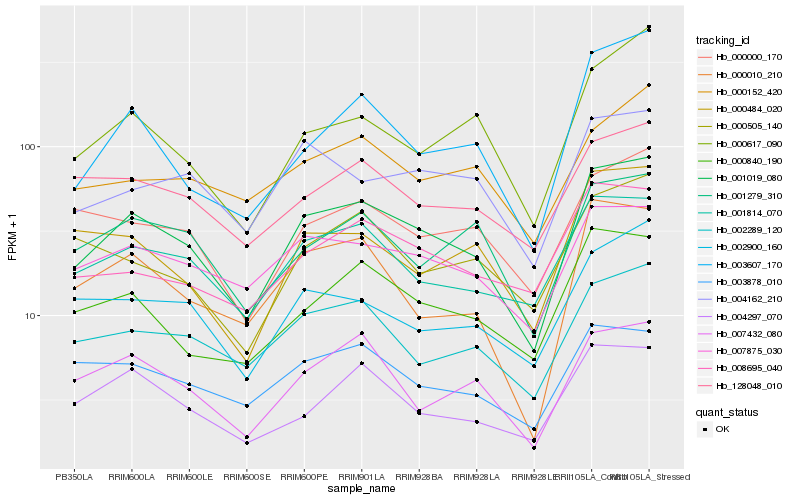
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 2 |
Hb_001814_070 |
0.1016827545 |
- |
- |
PREDICTED: uncharacterized protein LOC105650638 [Jatropha curcas] |
| 3 |
Hb_002289_120 |
0.1053470765 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000010_210 |
0.1088407995 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 5 |
Hb_000840_190 |
0.110193718 |
- |
- |
60S ribosomal protein L10, mitochondrial, putative [Ricinus communis] |
| 6 |
Hb_007432_080 |
0.110911537 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1-like [Jatropha curcas] |
| 7 |
Hb_007875_030 |
0.1111484352 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 8 |
Hb_001279_310 |
0.11365134 |
- |
- |
hypothetical protein JCGZ_10212 [Jatropha curcas] |
| 9 |
Hb_004162_210 |
0.1159208959 |
- |
- |
Cyclin-dependent kinases regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_000000_170 |
0.1164034576 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 11 |
Hb_004297_070 |
0.1165576241 |
- |
- |
hypothetical protein POPTR_0009s14820g [Populus trichocarpa] |
| 12 |
Hb_002900_160 |
0.1181188724 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 13 |
Hb_008695_040 |
0.1191061261 |
- |
- |
PREDICTED: small ribosomal subunit protein S13, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_128048_010 |
0.1215505129 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 15 |
Hb_003607_170 |
0.1219969065 |
- |
- |
U6 snRNA-associated Sm-like protein LSm5 [Hevea brasiliensis] |
| 16 |
Hb_003878_010 |
0.1228451313 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 17 |
Hb_000617_090 |
0.1241435471 |
- |
- |
hypoia-responsive family protein 4 [Hevea brasiliensis] |
| 18 |
Hb_000484_020 |
0.1248383948 |
- |
- |
hypothetical protein CISIN_1g030884mg [Citrus sinensis] |
| 19 |
Hb_000152_420 |
0.1257812216 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 20 |
Hb_000505_140 |
0.1264585096 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 3 [Populus euphratica] |